Abstract
Dementia is related to the cellular accumulation of β-amyloid plaques, tau aggregates, or α-synuclein aggregates, or to neurotransmitter deficiencies in the dopaminergic and cholinergic pathways. Cellular and neurochemical changes are both involved in dementia pathology. However, the role of dopaminergic and cholinergic networks in metabolic connectivity at different stages of dementia remains unclear. The altered network organisation of the human brain characteristic of many neuropsychiatric and neurodegenerative disorders can be detected using persistent homology network (PHN) analysis and algebraic topology. We used 18F-fluorodeoxyglucose positron emission tomography (18F-FDG PET) imaging data to construct dopaminergic and cholinergic metabolism networks, and used PHN analysis to track the evolution of these networks in patients with different stages of dementia. The sums of the network distances revealed significant differences between the network connectivity evident in the Alzheimer’s disease and mild cognitive impairment cohorts. A larger distance between brain regions can indicate poorer efficiency in the integration of information. PHN analysis revealed the structural properties of and changes in the dopaminergic and cholinergic metabolism networks in patients with different stages of dementia at a range of thresholds. This method was thus able to identify dysregulation of dopaminergic and cholinergic networks in the pathology of dementia.
Similar content being viewed by others
Introduction
Dementia is a neurodegenerative disease characterised by a progressive and chronic loss of function in cognitive, motor, sensory, and other brain systems1. The causes of Alzheimer’s disease (AD) and some other types of dementia are related to cellular pathologies, such as β-amyloid plaques2, tau aggregates3 and α-synuclein aggregates4 or to neurotransmitter deficiencies in the dopaminergic5 and cholinergic6 systems. Alterations in cholinergic circuity have been reported in disorders of attention and cognition7,8. Abnormal it contributes to memory and reward dysfunction in AD9. Dopamine appears to be particularly involved in the regulation of cognitive processes, and also has a functional relationship with the cholinergic system10. Furthermore, the accumulation of α-synuclein affects cholinergic and dopaminergic deficits.
Brain 18F-fluorodeoxyglucose positron emission tomography (18F-FDG PET) has been used for many years to diagnose dementia. The main advantage of 18F-FDG PET is its high sensitivity in detecting pathologies at the molecular level, due to the transportation of 18F-fluorodeoxyglucose into the intracellular space by glucose-1 transporters and its subsequent phosphorylation through the hexokinase reaction. 18F-FDG PET is used to measure energy consumption in neurons11, and it is associated with synaptic density and function. This method is sensitive to pathological changes in metabolic and biochemical processes, in contrast to magnetic resonance imaging, which can only visualise morphological changes. Using the 18F-FDG PET metabolic network, local neural activity, disconnection, and neuropathology effects can be observed. Each brain region can be assumed to exchange information directly or indirectly with other parts of the network through synchronized fluctuations in glucose uptake. Thus, we converted the brain images we obtained using 18F-FDG PET into a metabolic network and analysed the dopaminergic5 and cholinergic6 network connectivity between brain regions.
Network science involves measuring the similarity or difference between networks based on topology. Graph theory can be used to investigate the attributes of brain function and structure, and to do this the brain must be completely characterised as a network to understand brain function12. However, classical graph theory measures are usually based on either node or edge measurements, without considering aspects of network topology, such as connected components. Graph theory measures, such as the characteristic path length (CPL)13, network diameter (ND)14, and modularity (Mod)15,16, have been widely used to study the metabolic brain networks of patients with AD and mild cognitive impairment (MCI). These studies have reported topological alterations, resulting in lower efficiency, based on CPL, ND and disrupted Mod organisation17. The persistent homology network (PHN)18 is a mathematical tool in computational topology used for analysing the topological features of data that are persistent across in-network. PHN analysis was proposed as a precise and robust method for detecting differences in brain connectivity in AD19. It uses a topological feature (such as the number of connected brain regions)20 and a connected aggregation cost. This PHN feature is a monotonically decreasing convergence function and, when plotted across all possible filtration values, enables tracking network evolution between different groups. The connected component aggregation cost is derived from the minimum spanning tree of the network; thus, it can be considered the least efficient glucose metabolism pathway21 required for the evolution of the fully connected components.
The role of dopaminergic and cholinergic network connectivity in different stages of dementia is still unclear. PHN analysis can indicate how subcomponents of circuits are integrated into full connection when the filtration value increases. We studied changes in brain connectivity at different stages of dementia through the topological patterns of the dopaminergic and cholinergic networks derived from PHN analysis and successfully distinguished alterations in metabolic connectivity in the brain using 18F-FDG PET imaging. In this way we were able to identify dysregulation of dopaminergic and cholinergic networks in the pathology of dementia.
Results
We classified the values of the graph-based network indices CPL, ND, and eigenvector centrality (EC) into four groups based on their weighted networks after edge filtering; their corresponding P values passed the statistical threshold (10,000 permutations; P < 0.001). We also obtained the multinetwork according to graph filtration (λ) and computed four separate slopes of the integrated persistent plot of the area under the curve (SIP AUC) for patients with AD, MCI, and subjective cognitive decline (SCD), as well as for healthy controls (HC). All brain network parameters are presented in Supplementary Table S1.
Dopaminergic network analysis
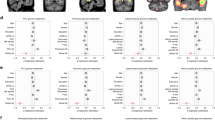
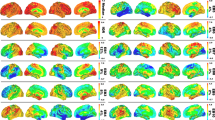
For the striatocortical pathways of the dopaminergic network, the single linkage distance (SLD) and dendrogram for each group are displayed in Fig. 1a,b. The SLD represents the functional distance between two brain regions. When λ increased, the dendrogram indicated that modularity occurred from the dorsal striatum to the prefrontal and sensorimotor cortices and that the supplementary motor regions had large delays in connectivity in patients with AD and MCI, but most markedly for MCI. The models showed a global increase in SLD for each connected region in the AD and MCI groups compared with the SCD and HC groups. To visualise network changes in the striatocortical pathways of the dopaminergic network during graph filtration, representative graphs are displayed at different filtration thresholds (λ = 0.05, 0.10, …, 0.25; Fig. 1c,d). When λ increased, the clustering of brain connections occurred more slowly and at a longer distance (less efficiency in the network) in the AD and MCI groups than in the HC and SCD groups. The pattern of delay was observed as follows: MCI > AD > SCD > HC. Moreover, we observed a significant difference between the CPL and SIP AUC values of various group pairs (Supplementary Table S1).
Dopaminergic network analysis. The dorsal dopamine pathway was affected in the Alzheimer’s disease (AD) and mild cognitive impairment (MCI) groups, showing a delay in connectivity and changes in configuration. (a) Single linkage distance (SLD) in the AD, MCI, subjective cognitive decline (SCD), and healthy control (HC) groups, obtained from the original correlation-based distance. (b) Single linkage dendrograms for the AD, MCI, SCD, and HC groups. The vertical and horizontal axes represent brain regions and graph filtration. The connected network is divided into smaller regions when the distance (graph filtration) decreases. (c) The corresponding persistent features of each group at different scales of graph filtration and (d) striatocortical network maps constructed using five different filtration values: 0.05, 0.10, 0.15, 0.20, and 0.25.
For the mesolimbic pathways of the dopaminergic network, the SLD and dendrogram for each group are displayed in Supplementary Figure S1a and b. The dendrograms revealed that for higher values of λ, clustering occurred later in the frontal lobe, amygdala, and anterior and posterior cingulate cortex than in other regions in the MCI group. Network changes in the mesolimbic pathways of the dopaminergic network are presented in Supplementary Figure S1c and d. For higher values of λ, the brain connection clustering was slower and less efficient in the AD and MCI groups than in the HC and SCD groups. No significant differences were noted between the SIP AUC values for the four groups, whereas a significant difference was noted between the CPL values for the MCI and SCD groups (Supplementary Table S1).
Cholinergic network analysis
For the Ch4 lateral capsular pathway of the cholinergic network, the SLD and dendrogram for each group are displayed in Fig. 2a,b. The dendrograms revealed that for higher values of λ, the distance in the temporal, frontal and parietal lobes was greater in the AD and MCI groups, especially the MCI group. In addition, the following delay pattern could be observed: MCI > AD > SCD > HC. To visualise network changes within the Ch4 lateral capsular pathway of the cholinergic network during graph filtration, representative graphs are displayed at various filtration thresholds (λ = 0.05, 0.10, …, 0.25; Fig. 2c,d). For higher values of λ, the brain connection clustering was less efficient and less dense in the AD and MCI groups than in the HC and SCD groups. We observed a significant difference in the SIP AUC for the Ch4 lateral capsular pathway network for the MCI and HC groups and for the MCI and SCD groups (Supplementary Table S1).
Cholinergic network analysis of the pathways of the Ch4 lateral capsular division reveal a greater distance in the temporal lobe, frontal lobe, and parietal lobe in the Alzheimer’s disease (AD) and mild cognitive impairment (MCI) groups, particularly in the MCI group. (a) Single linkage distance (SLD) in the AD, MCI, subjective cognitive decline (SCD), and healthy control (HC) groups, obtained from the original correlation-based distance. (b) Single linkage dendrograms for the AD, MCI, SCD, and HC groups. Vertical and horizontal axes represent brain regions and graph filtration, respectively. When the distance (graph filtration) decreases, the connected network is divided into smaller regions. (c) The corresponding persistent features of each group at different scales of graph filtration and (d) Ch4 lateral capsular projection network maps constructed using five different filtrations values: 0.05, 0.10, 0.15, 0.20, and 0.25.
For the Ch1–3 pathways of the cholinergic network, the SLD and dendrogram for each group are displayed in Supplementary Figure S2a and b. The dendrograms revealed that clustering occurred later and at a greater distance in the hippocampus and parahippocampus than in other regions in the AD and MCI groups. Network changes in the Ch1–3 pathways of the cholinergic network are displayed at various filtration thresholds in Supplementary Figure S2c and d. For higher values of λ, the brain region clustering was slower and less efficient in the AD and MCI groups than in the HC and SCD groups.
For the Ch4 medial pathway of the cholinergic network, the SLD and dendrogram for each group are displayed in Supplementary Figure S3a and b. The dendrograms revealed that clustering occurred later in the frontal gyrus than in other regions in the AD and MCI groups. The frontal regions were less connected with the cingulate cortex in the AD, MCI, and SCD groups. Network changes in the Ch4 medial pathway of the cholinergic network are displayed at various filtration thresholds in Supplementary Figure S3c and d. For higher values of λ, the brain connection clustering was sparser in the AD, MCI, and SCD groups.
For the Ch4 lateral perisylvian pathway of the cholinergic network, the SLD and dendrogram for each group are displayed in Supplementary Figure S4a and b. The dendrograms revealed a greater distance for the Herschel gyrus, insula and olfactory cortex than for the other regions in the AD and MCI groups. Network changes in the Ch4 lateral perisylvian pathway during graph filtration of the cholinergic network are displayed at various filtration thresholds (λ = 0.05, 0.10, …, 0.25) in Supplementary Figure S4c and d. For higher values of λ, brain connection clustering was slower and less efficient in the AD and MCI groups than in the HC and SCD groups.
In the Ch5–6 pathway of the cholinergic network, the thalamic and midbrain regions were the most affected in the AD and MCI groups. The SLD and dendrogram for each group are displayed in Supplementary Figure S5a and b. The dendrograms revealed that for higher values of λ, the distance in the ventral striatum and brainstem was greater in the AD and MCI groups. To visualise network changes during graph filtration of the Ch5–6 pathway in the cholinergic network, representative graphs are displayed at various filtration thresholds (λ = 0.05, 0.10, …, 0.25) in Supplementary Figure S5c and d. For higher values of λ, the brain connection clustering was less efficient and less dense in the AD and MCI groups than in the HC and SCD groups.
The network parameters of the cholinergic network were a few pairs showed a significant difference (Supplementary Table S1).
Discussion
In the metabolism network, each brain region exchanges information directly or indirectly with other parts of the network, as indicated by synchronized fluctuations in glucose uptake. We demonstrated that PHN analysis of 18F-FDG PET data for patients with different stages of dementia, with different levels of glucose metabolism connectivity, could reveal direct neurotransmitter networks and topological differences. The AD and MCI groups showed neurotransmitter network deficiencies in their dopaminergic and cholinergic networks and had a topological organisation indicating a less efficient topology structure. The PHN analysis revealed whether these brain regions remained intact or had been altered in each group. Importantly, these findings can locate the brain regions that change with disease progression. Ultimately, they may help in assessing the effects of treatments or other interventions on the brain. The hypothesized presence of differences in neurotransmitter networks between patients with MCI or AD and HCs or patients with SCD was confirmed.
The relationship between the dopaminergic network and amyloid-beta pathology is unclear22, and their involvement in AD is debatable23,24. In general, the release of dopamine transmitter in the caudate, putamen, hippocampus, and frontal cortex of the brain decreases during ageing25. The network containing the striatocortical pathways (projections to the sensory–motor cortex) is involved in controlling voluntary movements. The prefrontal cortices are implicated in executive function, and distinct areas of the prefrontal cortex have strong functional connections with striatocortical pathways26. The mesolimbic pathway, targeting the hippocampi, cerebral cortex and nucleus accumbens, is responsible for cognitive and behavioural signs. Both the prefrontal cortices and the mesolimbic pathway may be involved in AD progression5. The decline of or a deficit in the dopaminergic pathway is predictive of ‘unsuccessful ageing’ and is considered an indicator of frailty in older adults27. The CPL and SIP AUC are directly related to the local topology and efficiency of the network. We observed significant differences in the CPL and SIP AUC among the groups. The AD and MCI groups showed slower network integration rates and lower efficiency than the SCD and HC groups in executive and motor functions of the striatocortical pathway network. Some studies have shown strong correlations between dopamine depletion in the striatocortical pathway and deficits in executive function, as indicated using the object alternation task28 and the Stroop test29. In addition, other studies have indicated that the dopamine and choline systems are tightly interconnected and involved in various cognitive functions, such as memory, attention, and learning30,31,32.
The limbic and paralimbic cortices of the brain receive the heaviest cholinergic input from Ch4 and are also the main sources of mutual projections back to the nucleus basalis. This limbic affiliation explains the role of the basal nucleus in regulating the influence of incoming sensory information and memory. Several studies have reported that AD affects cholinergic system degeneration33,34. Cholinergic hypofunction is related to memory deficit progression with ageing. Normal ageing is accompanied by a gradual loss of cholinergic function. Our results showed significant differences in the CPL and SIP AUC of the Ch4 lateral capsular pathway of the cholinergic network among the dementia groups: the AD and MCI groups had slower network integration and lower efficiency in the subbranches of the cholinergic network than the SCD and HC groups. This pathology in Ch4 eventually leads to the degeneration of the part of the cholinergic network, which Ch4 sends to the cerebral cortex. The early involvement of Ch4 has an amplifying effect on Alzheimer’s pathology, because degeneration of Ch4 can perturb neurotransmission in all cortical areas35. Our results are consistent with the idea that the cholinergic network has a high demand for energy production and therefore should respond more sensitively to ageing-related energy (glucose) deprivation36. According to our PHN analysis results, the MCI group had slower process and sparser connectivity than the AD group; the MCI group may thus have an increased risk of AD (approximately 20% conversion)37,38. Cholinergic dysfunction plays a key role in the clinical development of MCI. Previous observations have suggested that disruptions in the metabolic processes of the cholinergic system lead to the full dementia deficits seen in AD, and that this disruption can be an early marker of disease progression39. Dysfunction of the cholinergic system can be treated pharmacologically with cholinergic medication, which can ameliorate the cholinergic deficit during the early stages of the disease and retard disease progression.
This study has some strengths and limitations. Traditional graph theory analysis identifies one fixed threshold, which may or may not completely characterise the brain structure network. In contrast, PHN analysis considers topological changes at all possible thresholds, which provides a more complete characterisation of the brain network. In this study, we showed that in a persistent homology context, network evolution can be represented by bar codes. The anatomical regions were based on neurotransmitter mapping and histopathology, however only anatomical regions specifically constructed for brain systems were used.
In summary, our study results demonstrated pattern topology alterations in the dopaminergic and cholinergic networks at different stages of dementia. MCI and AD could be associated with disease progression. The PHN analysis demonstrated the integration of subregions in the network into full connection at different scales and distinguished alterations in brain 18F-FDG PET metabolic connectivity as a model of dementia. We identified the dysregulation of the dopaminergic and cholinergic networks in the pathology of dementia.
Methods
Participants
We recruited 73 individuals and obtained 18FDG-PET data for them. The clinical evaluation performed in this study revealed that 16 individuals had early AD, 18 had MCI, 16 had SCD, and 23 were HCs. The detailed cohort information and Mini-Mental State Examination results40 are presented in Table 1. Age, sex, height, weight, and body mass index did not differ significantly among the four groups. An AD diagnosis was established during a multidisciplinary consensus meeting, according to the clinical criteria for probable AD from the National Institute on Aging–Alzheimer’s Association41. MCI was diagnosed according to the revised consensus criteria from 200442. The criteria for SCD in pre-MCI were based on a previous study43. All participants or their caregivers provided written informed consent for the experimental procedures, which were approved by the ethics committee of the Taipei Veterans General Hospital. All methods were carried out in accordance with the relevant guidelines and regulations of this ethics committee. The protocol conformed to the ethical standards of the Declaration of Helsinki for the protection of human participants.
MR and 18FDG-PET data acquisition and preprocessing
Metabolic and structural images were obtained using a hybrid 3 T Signa PET/MR scanner (GE Healthcare, Milwaukee, WI, USA), which could simultaneously acquire both time-of-flight PET and high-resolution magnetic resonance (MR) data. All participants fasted for six hours before the 18F-FDG PET injection (~ 185 MBq), and all PET scans started approximately 60 min after the injection. T1-weighted images were obtained using the 3D BRAVO sequence (TR = 8 ms; TE = 3 ms; FA = 12°; FOV = 25.6; and 192 slices with a thickness of 1 mm). The T1-weighed structural images were processed using the CAT 12 toolbox44 in Matlab (MathWorks, Natick, MA, USA) and mapping to the Montreal Neurological Institute (MNI) template. The brain structural data were then segmented into areas of grey matter, white matter, and cerebrospinal fluid. Bias correction and modulation were performed to remove intensity nonuniformities in the grey matter. Finally, all the images were smoothed using a Gaussian filter (8 mm full width at half maximum [FWHM]).
The 18F-FDG PET images were co-registered with the T1‐weighted MR images and then spatially normalised to the MNI atlas using the software toolkit Statistical Parametric Mapping (SPM12, Wellcome Department of Cognitive Neurology, London, UK) in Matlab. The normalised 18F-FDG PET images were corrected for partial volume effects and the PETPVE12 toolbox was applied45. Subsequently, the standardised uptake values of the 18F-FDG PET images were normalised to the mean value for the cerebellum. We conducted spatial smoothing with a Gaussian kernel for the FWHM equal to (8,8,8) in three directions (x,y,z) to improve the signal-to-noise ratio.
Node selection and network construction
We focused on the neurotransmitter networks related to dementia pathology and selected the cholinergic and dopaminergic neurotransmission pathways. We created each module of the metabolic connectivity network according to the method proposed by Caminiti et al.46.
For the dopaminergic network analysis, we created striatocortical and mesolimbic networks (Fig. 3). The striatocortical network consisted of the dorsal caudate and dorsal putamen, frontal premotor, motor, executive dorsolateral frontal regions, and somatosensory cortex. The mesolimbic network consisted of the ventral striatum, ventral and medial frontal areas, anterior and middle cingulate cortices, amygdala, and parahippocampal cortex.
For the cholinergic network analysis, we created five cholinergic networks (Fig. 4). The first network consisted of brain regions innervated by Ch1, including the Ch2 nuclei of the forebrain, namely the bilateral hippocampus and hypothalamus, and the pathway arising from the Ch3 nucleus, which includes the olfactory and parahippocampal cortices.
The cholinergic neurons in the brainstem pathways are the Ch5 pedunculopontine tegmental nucleus and the Ch6 laterodorsal tegmental grey of the periventricular area, which extend to the thalamus, ventral and dorsal striatum, globus pallidus, and the brainstem reticular formation, which is represented by the pons, midbrain, and medulla oblongata.
The other three cholinergic networks were represented by pathways originating from the Ch4 nuclei. The first Ch4 medial pathway reaches the cingulate, retrosplenial, and orbitofrontal cortices. The second Ch4 lateral perisylvian division joins the olfactory and superior temporal cortices with the insula and frontoparietal operculum. The third Ch4 lateral capsular division connects to the remaining frontal, parietal, temporal, and occipital cortices, as well as the amygdala.
We created dopaminergic and cholinergic subject-by-node/region-of-interest (ROI) matrices for each group. The matrices contained regional cerebral metabolic values derived from each subject in each node/ROI.
PHN analysis
A flowchart summarising the PHN analysis methodology is presented in Fig. 5. Each group of preprocessed 18F-FDG PET images had ROIs extracted from the cholinergic and dopaminergic networks, and Pearson’s correlation coefficients between intra-networks were calculated across individuals. To calculate the PHN, the distances between regions were measured. We converted the correlation-based matrix to a distance matrix using the formula 1 −|correlation|. The PHN enabled us to quantify various persistent topological features based on graph filtration (λ). Using zeroth Betti numbers to create a persistent homology graph of filtration enabled us to track the construction of all regions connected to the network as the filtration value increased47. During this construction, we could record the start, the time at which a region appeared, and the end for various filtration values. With respect to filtration, when λ = 0, all regional components are loose: the number of connected components decreases with an increase in filtration. We can use the SIP as a rate of component convergence. However, the slope is not the most appropriate method to describe the plot, since it is nonlinear. We considered the AUC of the SIP to represent the sum of the distances of all paths. The AUC of the SIP is equal to efficient glucose metabolism demand within the network due to weak and strong connections.
Flowchart of the persistent homology network (PHN) analysis methodology. Each group of preprocessed 18F-FDG PET images had regions of interest extracted within networks, calculated using Pearson’s correlation coefficients between intra-networks and across individuals. We converted the correlation-based matrix to a distance matrix using the formula 1—|correlation|. Using zeroth Betti numbers to create a persistent homology graph of filtration enabled us to track the construction of all regions connected to the network as the filtration value increased.
Each topological feature was displayed using a bar that started and ended when the feature appeared and disappeared, respectively. This bar code was used to quantify and visualise evolutionary changes in topological features that occurred when the graph filtration changed. By incorporating additional geometric information into the bar code, we obtained an SLD and a dendrogram that showed the overall evolution and the hierarchical connections between the components of the network. We visualised these changes during filtration using the bar codes and dendrograms. The bar codes decreased with the threshold for ‘the number of regions’ to 1. If the network was strongly connected at a small threshold of distance (dij), the SIP decreased rapidly.
Graph theory analysis
To identify the persistent homology in the cholinergic and dopaminergic networks, the network must be represented as a simplicial complex. A distance function in the underlying network corresponds to a filtration of the simplicial complex. Graph theory analysis has three other graph-theoretical parameters related to the distance function to estimate intra-network parameters: CPL, ND, and EC. These parameters were obtained using the Brain Connectivity Toolbox, which is an open-source Matlab code (http://www.brain-connectivity-toolbox.net). CPL is considered the average shortest distance between all regional pairs13. For example, a low CPL indicates a network with easy or quick data transfer. ND indicates how far the furthest regions of a network are from one another, based on paired path lengths. It provides an understanding of the network size. A high ND and low CPL would, therefore, be considered to indicate an efficient network. EC assigns increased importance to regions that are connected to other highly connected regions through the intra-network48.
Statistical analysis
Data analyses were conducted in two steps: demographic and main analyses. The demographic analysis compared demographic variables with the clinical characteristics of the four groups of patients (AD, MCI, SCD and HC) to identify any potentially confounding relationships. The demographic analyses were performed using Mann–Whitney U and chi-square tests to identify differences among the groups. We used SPSS 13 software (SPSS, Chicago, IL, USA) for the analyses, with statistical significance assumed at P < 0.0549. To obtain stable results in the main analysis of the network parameters, we used a permutation test considering 10,000 permutations between any two groups in addition to significant differences between any pair of groups at a significance level of 0.001. This statistical method has been described in a previous study50.
References
Epstein, F. H. & Martin, J. B. Molecular basis of the neurodegenerative disorders. New Engl. J. Med. 340, 1970–1980 (1999).
Cummings, B. J. & Cotman, C. W. Image analysis of β-amyloid load in Alzheimer’s disease and relation to dementia severity. Lancet 346, 1524–1528 (1995).
Mandelkow, E.-M. & Mandelkow, E. Tau in Alzheimer’s disease. Trends Cell Biol. 8, 425–427 (1998).
Lippa, C. F. et al. Lewy bodies contain altered α-synuclein in brains of many familial Alzheimer’s disease patients with mutations in presenilin and amyloid precursor protein genes. Am. J. Pathol. 153, 1365–1370 (1998).
Martorana, A. & Koch, G. Is dopamine involved in Alzheimer’s disease?. Front. Aging Neurosci. 6, 252 (2014).
Mesulam, M. The cholinergic lesion of alzheimer’s disease: Pivotal factor or side show?. Learn Mem. 11, 43–49 (2004).
Wallace, T. L., Ballard, T. M., Pouzet, B., Riedel, W. J. & Wettstein, J. G. Drug targets for cognitive enhancement in neuropsychiatric disorders. Pharmacol. Biochem. Behav. 99, 130–145 (2011).
Higley, M. J. & Picciotto, M. R. Neuromodulation by acetylcholine: Examples from schizophrenia and depression. Curr. Opin. Neurobiol. 29, 88–95 (2014).
Nobili, A. et al. Dopamine neuronal loss contributes to memory and reward dysfunction in a model of Alzheimer’s disease. Nat. Commun. 8, 14727 (2017).
Martorana, A. et al. Dopamine modulates cholinergic cortical excitability in Alzheimer’s disease patients. Neuropsychopharmacology 34, 2323–2328 (2009).
Magistretti, P. J., Pellerin, L., Rothman, D. L. & Shulman, R. G. Neuroscience: Energy on demand. Science 283, 496–497 (1999).
Sporns, O., Tononi, G. & Kötter, R. The human connectome: A structural description of the human brain. PLoS Comput. Biol. 1, e42 (2005).
Brier, M. R. et al. Functional connectivity and graph theory in preclinical Alzheimer’s disease. Neurobiol. Aging 35, 757–768 (2013).
Fagerholm, E. D., Hellyer, P. J., Scott, G., Leech, R. & Sharp, D. J. Disconnection of network hubs and cognitive impairment after traumatic brain injury. Brain J. Neurol. 138, 1696–1709 (2015).
Rubinov, M. & Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 52, 1059–1069 (2009).
Sporns, O. & Betzel, R. F. Modular brain networks. Annu. Rev. Psychol. 67, 613–640 (2015).
de Haan, W. et al. Disrupted modular brain dynamics reflect cognitive dysfunction in Alzheimer’s disease. Neuroimage 59, 3085–3093 (2012).
Aktas, M. E., Akbas, E. & Fatmaoui, A. E. Persistence homology of networks: Methods and applications. Appl. Netw. Sci. 4, 61 (2019).
Kuang, L. et al. A concise and persistent feature to study brain resting-state network dynamics: Findings from the Alzheimer’s Disease Neuroimaging Initiative. Hum. Brain Mapp. 40, 1062–1081 (2018).
Lee, H., Kang, H., Chung, M. K., Kim, B.-N. & Lee, D. S. Persistent brain network homology from the perspective of dendrogram. IEEE Trans. Med. Imaging 31, 2267–2277 (2012).
Shokri-Kojori, E. et al. Correspondence between cerebral glucose metabolism and BOLD reveals relative power and cost in human brain. Nat. Commun. 10, 690 (2019).
Kar, S., Slowikowski, S. P. M., Westaway, D. & Mount, H. T. J. Interactions between beta-amyloid and central cholinergic neurons: implications for Alzheimer’s disease. J. Psychiatry Neurosci. Jpn. 29, 427–441 (2004).
Trillo, L. et al. Ascending monoaminergic systems alterations in Alzheimer’s disease. Translating basic science into clinical care. Neurosci. Biobehav. Rev. 37, 1363–1379 (2013).
Portet, F., Scarmeas, N., Cosentino, S., Helzner, E. P. & Stern, Y. Extrapyramidal signs before and after diagnosis of incident Alzheimer disease in a prospective population study. Arch. Neurol.-chicago 66, 1120–1126 (2009).
Bäckman, L., Lindenberger, U., Li, S.-C. & Nyberg, L. Linking cognitive aging to alterations in dopamine neurotransmitter functioning: recent data and future avenues. Neurosci. Biobehav. R 34, 670–677 (2009).
Fuster, J. M. The Prefrontal Cortex (Fourth Edition). 221–283 (2008) https://doi.org/10.1016/b978-0-12-373644-4.00006-2.
Depp, C. A. & Jeste, D. V. Definitions and predictors of successful aging: A comprehensive review of larger quantitative studies. Am. J. Geriatr. Psychiatry 14, 6–20 (2006).
Marié, R. M. et al. Relationships between striatal dopamine denervation and frontal executive tests in Parkinson’s disease. Neurosci. Lett. 260, 77–80 (1999).
Brück, A. et al. Positron emission tomography shows that impaired frontal lobe functioning in Parkinson’s disease is related to dopaminergic hypofunction in the caudate nucleus. Neurosci. Lett. 311, 81–84 (2001).
Cara, B. D. et al. Activation of dopamine D1 receptors enhances cholinergic transmission and social cognition: A parallel dialysis and behavioural study in rats. Int. J. Neuropsychopharmacol. 10, 383 (2006).
Cao, Y.-J., Surowy, C. S. & Puttfarcken, P. S. Different nicotinic acetylcholine receptor subtypes mediating striatal and prefrontal cortical [3H]dopamine release. Neuropharmacology 48, 72–79 (2005).
Millan, M. J. et al. Selective blockade of dopamine D 3 versus D 2 receptors enhances frontocortical cholinergic transmission and social memory in rats: a parallel neurochemical and behavioural analysis. J. Neurochem. 100, 1047–1061 (2007).
Geula, C., Nagykery, N., Nicholas, A. & Wu, C.-K. Cholinergic neuronal and axonal abnormalities are present early in aging and in Alzheimer disease. J. Neuropath Exp. Neurol. 67, 309–318 (2008).
Gil-Bea, F. J., García-Alloza, M., Domínguez, J., Marcos, B. & Ramírez, M. J. Evaluation of cholinergic markers in Alzheimer’s disease and in a model of cholinergic deficit. Neurosci. Lett. 375, 37–41 (2005).
Mesulam, M.-M. Cholinergic circuitry of the human nucleus basalis and its fate in Alzheimer’s disease. J. Comp. Neurol. 521, 4124–4144 (2013).
Szutowicz, A. et al. Phenotype-dependent susceptibility of cholinergic neuroblastoma cells to neurotoxic inputs. Metab. Brain Dis. 21, 143–155 (2006).
Fischer, P. et al. Conversion from subtypes of mild cognitive impairment to Alzheimer dementia. Neurology 68, 288–291 (2007).
Maioli, F. et al. Conversion of mild cognitive impairment to dementia in elderly subjects: A preliminary study in a memory and cognitive disorder unit. Arch. Gerontol. Geriatr. 44, 233–241 (2007).
Marcone, A. et al. [11C]-MP4A PET cholinergic measurements in amnestic mild cognitive impairment, probable Alzheimer’s disease, and dementia with Lewy bodies: a Bayesian method and voxel-based analysis. J. Alzheimer’s Dis. 31, 387–399 (2012).
Folstein, M. F., Folstein, S. E. & McHugh, P. R. Mini-mental state. J. Psychiatr. Res. 12, 189–198 (1975).
McKhann, G. M. et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dementia J. Alzheimer’s Assoc. 7, 263–269 (2011).
Petersen, R. C. Mild cognitive impairment as a diagnostic entity. J. Intern. Med. 256, 183–194 (2004).
Jessen, F. et al. A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer’s disease. Alzheimer’s Dementia 10, 844–852 (2014).
Gaser, C. & Dahnke, D. CAT-a computational anatomy toolbox for the analysis of structural MRI data. HBM (2016).
Gonzalez-Escamilla, G. et al. PETPVE12: An SPM toolbox for partial volume effects correction in brain PET—Application to amyloid imaging with AV45-PET. Neuroimage 147, 669–677 (2017).
Caminiti, S. P. et al. Metabolic connectomics targeting brain pathology in dementia with Lewy bodies. J. Cereb. Blood Flow Metab. 37, 1311–1325 (2016).
Giusti, C., Ghrist, R. & Bassett, D. S. Two’s company, three (or more) is a simplex: Algebraic-topological tools for understanding higher-order structure in neural data. J. Comput. Neurosci. 41, 1–14 (2016).
van Duinkerken, E. et al. Altered eigenvector centrality is related to local resting-state network functional connectivity in patients with longstanding type 1 diabetes mellitus. Hum. Brain Mapp. 38, 3623–3636 (2017).
Lin, W.-C. et al. Reestablishing brain networks in patients without overt hepatic encephalopathy after liver transplantation. J. Cereb. Blood Flow Metab. 34, 1877–1886 (2014).
Bernhardt, B. C., Chen, Z., He, Y., Evans, A. C. & Bernasconi, N. Graph-theoretical analysis reveals disrupted small-world organization of cortical thickness correlation networks in temporal lobe epilepsy. Cereb. Cortex 21, 2147–2157 (2011).
Acknowledgements
The study was supported by grants from the Academia Sinica of Taiwan (AS-BD-108-2), the Ministry of Science and Technology of Taiwan (107-2221-E-075-006- and 109-2314-B-075-052-MY2), the Taipei Veterans General Hospital (V108C-113, VGHUST109-V1-5-1, and V109C-061), the Wong Vung-Hau Radiology Foundation, and the Brain Research Center, National Yang-Ming University, from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. We thank our colleagues from the Digital Medicine Initiative. The Institute of Information Science, Academia Sinica of Taiwan provided insight and expertise that greatly assisted this research, although the interpretations and conclusions presented in this paper are not necessarily representative of their views. We would like to thank Uni-edit (www.uni-edit.net) for editing and proofreading this manuscript.
Funding
The study was supported by grants from the Academia Sinica of Taiwan (AS-BD-108-2), the Ministry of Science and Technology of Taiwan (107-2221-E-075-006, 109-2314-B-075-052-MY2), the Taipei Veterans General Hospital (V108C-113, VGHUST109-V1-5-1, V109C-061), and the Brain Research Center, National Yang-Ming University from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Author information
Authors and Affiliations
Contributions
J.-L.F. and T.-W.H. conceived the idea for the study. T.-W.H. and C.-J.C. developed the theory, collected the data, and performed the computational analyses. D.-W.W. and L.-F.C. verified the analytical methods. W.-S.H., H.-M.W., and W.-Y.G. investigated and supervised the findings of this study. All authors discussed the results and contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hsu, TW., Fuh, JL., Wang, DW. et al. Disrupted metabolic connectivity in dopaminergic and cholinergic networks at different stages of dementia from 18F-FDG PET brain persistent homology network. Sci Rep 11, 5396 (2021). https://doi.org/10.1038/s41598-021-84722-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-84722-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.