« Prev Next »
The two X chromosomes in mammalian females function very differently. The X chromosome contains over 1,000 genes, while the Y chromosome is relatively gene poor. This presents a challenge to placental mammals, who need to (roughly) equalize expression from the X chromosome in male (XY) and female (XX) cells. The mammalian solution is therefore to inactivate one X chromosome in females during early development by assembling a distinctive form of chromatin that largely silences gene expression. Since the first pictures of the inactive X chromosome were published in 1949, scientists have studied the inactivation process in detail, and they have discovered that the X chromosome possesses a unique regulatory region, called the X-inactivation center, that is essential for inactivation.
Inactive X Chromosomes Can Be Detected in Interphase Cells
It is not difficult to pick out the inactive X chromosome in an interphase cell. Unlike other chromosomes, the inactive X never fully decondenses during interphase. Thus, the inactive X appears as a densely stained structure within the interphase nucleus of female cells. Murray Barr and Ewart Bertram provided some of the first images of the inactive X in a 1949 study, in which they noted the following:
"The sex of a somatic cell as highly differentiated as a neurone may be detected with no more elaborate equipment than a compound microscope following staining of the tissue by the routine Nissl method... The difference in nuclear structure between neurones of adult male and female cats rests on the degree of development of a second body, which is much smaller than the nucleolus."
Barr and Bertram called this second body, which we now refer to as a Barr body, a nucleolar satellite, because it was frequently located next to the nucleolus. Their data showed that the Barr body was well-developed in females (Figure 1) but poorly developed in males (Figure 2). Thus, the duo hypothesized that "the nucleolar satellite may be derived from the heterochromatin of the sex chromosomes." Researchers now appreciate how prescient these observations were, even though Barr and Bertram were unable to recognize that the satellite was a single X chromosome.
Ten years later, Susumu Ohno and colleagues (Ohno et al., 1959) provided more definitive evidence that the Barr body was an X chromosome. By systematically comparing female and male liver cells at various points in the cell cycle, they observed that female cells contained a highly condensed chromosome that was absent in chromosome spreads of male cells. Furthermore, they noted that triploid cells had additional copies of this condensed structure. From the data, Ohno et al. deduced that the Barr body, or sex chromatin, was likely a condensed X chromosome. Definitive proof of its identity would come later, after scientists developed molecular probes that could be used to identify specific chromosomes by in situ hybridization.
Lyon's X-Inactivation Hypothesis Bridges Genetics and Cytology
In the early 1960s, researcher Mary Lyon built on Ohno's cytological observations when she formulated the X-inactivation hypothesis (Lyon 1961, 1962). Lyon was a mouse geneticist who was intrigued by the fact that female, but not male, mice had a mottled coat color when they were heterozygous for the sex-linked genes involved in coat color. At the time, several X-linked genes had been implicated in coat color, and investigators were able to perform the breeding experiments that were necessary to understand the inheritance of coat color. Bringing together the genetic and cytological data, Lyon interpreted "sex chromatin" to be a manifestation of a genetically inactive X chromosome. She could then explain the mottled appearance of female mice as follows:
"It is here suggested that this mosaic phenotype is due to the inactivation of one or the other X chromosome early in embryonic development. If this is true, pigment cells descended from cells in which the chromosome carrying the mutant gene was inactivated will give rise to a normal-coloured patch and those in which the chromosome carrying the normal gene was inactivated will give rise to a mutant-coloured patch" (Lyon, 1961).
An essential element of Lyon's hypothesis was the random nature of the inactivation process, which could therefore affect either the maternally inherited or the paternally inherited X chromosome. Inactivation also explained why XO female mice were able to survive with a single X chromosome. In her second paper on this subject (1962), Lyon extended her analysis to other species, including cats and humans, concluding that X inactivation applies generally to mammals.
An X-Inactivation Center Is Located on the Long Arm of the X Chromosome
In the decades after Lyon proposed X inactivation, researchers produced a large body of data that was consistent with the theory, but the factors that controlled inactivation remained elusive. Cytogenetic studies of mouse and human cells with deleted or rearranged X chromosomes pointed to the existence of a unique region of the X chromosome, called the X-inactivation center (abbreviated XIC or Xic in humans and mice, respectively), which was crucial for the X-inactivation process to occur (Rastan & Robertson, 1985). By comparing a series of deletion chromosomes, investigators were able to narrow down the location of the XIC to a portion of the q arm of the X chromosome. A key element in the eventual mapping of the XIC was the availability of rodent-human hybrid cell lines that contained altered human X chromosomes. These lines had been derived by fusing human cells, obtained from patients who carried various X chromosome translocations or deletions, with rodent cells in tissue culture. The hybrid cell lines derived by this procedure generally contained only a few human chromosomes, which could be identified using chromosome-specific markers.
Using this method, Brown and colleagues (Brown et al., 1991a) studied hybrid cell lines that contained partial segments of the human X chromosome, asking whether any specific region of the X chromosome was required for inactivation. An inactivated X chromosome could be distinguished from other chromosomes in a cell by its relatively late replication during S phase, which is a characteristic feature of heterochromatin. Figure 3 shows idiograms of the rearranged human X chromosomes from four cell lines, compared to a normal X chromosome on the left. From this figure, taken from Brown et al.'s paper, it is clear that the altered X chromosomes have extensive rearrangements, and that there is little commonality between them. Nonetheless, all of the altered X chromosomes were still subject to inactivation. By careful observation of the breakpoints that had occurred during the original translocation or deletion event, Brown and colleagues were able to identify a common region on the long arm of the X chromosome that was present in all chromosomes that underwent inactivation. They then used in situ hybridization with a series of X chromosome DNA probes to further define the boundaries of the common region, which corresponded to the distal portion of region Xq13. The homologous region, Xic, had been identified in the mouse, and scientists subsequently used transgenic mouse models to establish the requirement for this region in X inactivation.
XIST RNA Is Produced from the Inactive X Chromosome
Quite serendipitously, Brown et al. also discovered an unusual transcript from the XIC region. In particular, while trying to clone another gene on the X chromosome, they discovered a gene that was specifically expressed from the XIC region on chromosomes that underwent inactivation (Brown et al., 1991a, b). Based upon this novel property, they named the gene XIST, for "X inactive specific transcript." To demonstrate that the XIST gene was expressed specifically from the inactive X, the researchers made a quantitative comparison of XIST transcription in normal and aneuploid human cell lines, as well as in rodent-human cell lines. Figure 4 shows the data from the experiment, which used DNA hybridization to measure the relative amount of XIST RNA in samples of total RNA prepared from the various cell lines. In this experiment, the intensity of a band is roughly proportionate to the amount of XIST RNA in the sample. The source of the RNA sample is indicated on the right. These data show clearly that XIST is expressed in normal (46,XX) females, but not in normal males (46,XY). In aneuploid cell lines with 47 to 49 chromosomes, the quantity of XIST transcripts is proportional to the number of supernumerary X chromosomes. The rodent-human hybrid cell lines were divided into Xa and Xi classes, reflecting whether the human X had been shown to be active or inactive, respectively. From the results, it is clear that XIST RNA is produced only from the inactive X and not from the active X in rodent-human hybrid cells. A control experiment showed that Xist transcripts are not produced from mouse chromosomes in the hybrid lines. (Human XIST corresponds to mouse Xist.) Taken together, the results show clearly that XIST is transcribed from inactivated X chromosomes.
XIST Transcripts Are Cis-Acting RNAs That Coat the Inactive X Chromosome
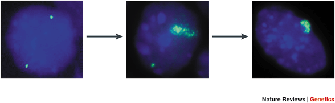
From the initial discovery of XIST, scientists suspected that XIST transcripts had unusual properties. Brown et al. had observed that multiple XIST transcripts were produced by alternative splicing in cells. However, there were many stop codons in all potential reading frames, making it improbable that XIST transcripts were translated in cells. Indeed, localization of XIST transcripts in cells by fluorescence in situ hybridization (FISH) confirmed these suspicions and pointed to a unique function of XIST transcripts. Clemson and colleagues (1996) noted that, unlike the transcripts of other single-copy genes, XIST transcripts formed a diffuse cluster that remained in the nucleus of normal female human fibroblasts. The number of XIST clusters appeared to be the same as the number of inactive X chromosomes, suggesting that XIST transcripts remained physically associated with the inactive X chromosome. This hypothesis was confirmed when the investigators simultaneously hybridized XIST probes with a collection of probes specific for the X chromosome. XIST clusters always co-localized with the X chromosome probes, which also gave a clustered appearance. Moreover, in aneuploid female cells, the number of XIST clusters was always one less than the number of X chromosome clusters, consistent with the presence of a single active X chromosome. Thus, the investigators concluded that XIST transcripts "painted" inactive X chromosomes along their length. They also noted that XIST did not interact directly with X chromosome DNA, because it was not removed by enzymatic degradation of the DNA. Based on these findings, they proposed that XIST transcripts play an architectural role in defining the nuclear territory of the inactive X chromosome, which fits well with current knowledge about the unique chromatin of the inactive X.
To define the role of XIST RNA in X inactivation, researchers needed an experimental model that could be genetically manipulated. Therefore, experimental work with X inactivation has been performed almost entirely with the laboratory mouse. Mouse embryonic stem (ES) cells provided a valuable model for studying the initial events in X inactivation (Martin et al., 1978), which occur in the early mammalian embryo. ES cells are derived from the inner cell mass of early embryos, and these cells remain pluripotent until they are induced to differentiate, after which X chromosomes undergo random inactivation. The presence of Xist transcripts in ES cells can be visualized by fluorescence in situ hybridization (FISH). Figure 5 documents the changes in Xist transcripts that accompany ES cell differentiation. The left panel shows an undifferentiated ES cell in which Xist transcription can be detected on both the maternal and paternal X chromosomes. At intermediate stages of differentiation (middle panel), transcription from both X chromosomes is still occurring, but Xist transcripts coat one X chromosome selectively. Then, when the ES cells are fully differentiated (right panel), Xist transcripts coat the inactive X, and Xist transcription is no longer detected from the other chromosome. Thus, the timing of Xist transcription is consistent with a fundamental role in cis-inactivation, but other questions arise. Why, for example, are Xist transcripts no longer detected on the active X after the cells have differentiated? And how do cells determine which X chromosome to inactivate? It turns out that Xist is only one activity encoded by the Xic. We now know that the Xic is a complex locus that includes other sequences involved in counting X chromosomes and determining which chromosome to inactivate.
Gene Knockouts Establish That Xist Is Required for Cis-Inactivation

The timing of Xist transcription and coating of inactive X chromosomes were consistent with, but could not prove, a requirement for Xist in X inactivation. To determine whether Xist is indeed required for X inactivation, researchers developed mouse models in which the normal Xist gene was inactivated. Penny et al. (1996) constructed female ES cell lines in which the Xist gene was deleted on one X chromosome. These particular ES cells were polymorphic for multiple X-linked genes, so the investigators were able to identify the specific chromosome that became inactivated when the cells differentiated. By using allele-specific probes for X-linked transcripts, researchers found that the chromosome with a functional Xist gene was always inactivated as the cells differentiated. Chromosomes with mutant Xist genes were no longer subject to inactivation. Thus, inactivation of an X chromosome requires a functional Xist gene on the same chromosome.
The physiological consequences associated with loss of Xist function were further explored by Marahrens et al. (1997), who generated knockout mice deficient in Xist activity. Interesting differences were observed between female and male mice in these experiments, consistent with the importance of X inactivation in early development. Not surprisingly, male mice were phenotypically normal, because X inactivation does not occur in most male tissues. The only exception is the male germ line, so researchers took note that males were fertile and produced sperm that were capable of fertilizing eggs. Two quite different results were observed in female mice, depending on whether the nonfunctional Xist gene was inherited from their mother or their father. Females who inherited the defective Xist from their mothers were phenotypically normal, and maternal-specific alleles were expressed in all tissues, indicating that the paternal X had been inactivated. Females who inherited the defective Xist from their fathers, however, were developmentally retarded and died as embryos. Expression analysis showed that the maternal X was inactivated in all of the embryonic cells, but that neither X chromosome was inactivated in the trophoblast, the extra-embryonic tissue that gives rise to the placenta. The lethality was attributed to the lack of dosage compensation in the trophoblast, which is where the paternal X is normally inactivated.
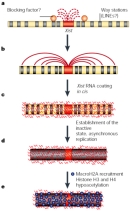
The cumulative results from these and many other experiments have established that Xist is not only necessary but also sufficient for cis-inactivation of the X chromosome. We now know, however, that Xist does not control all of the functions attributed to the Xic. Other sequences within the Xic are important for counting the number of X chromosomes and selecting the X to be inactivated. Figure 6 summarizes elements and regions of the Xic that have been implicated in its choice, counting, and cis-inactivation functions. (The positions of the five deletions that were used to map these functions are shown by black lines 1-5.) Note that the Xist locus occupies only a small portion of the much larger Xic, which has been functionally mapped to a 450-kilobase segment of the X chromosome. One feature of particular interest is the initiation site for the 40-kilobase antisense transcript Tsix, which is expressed from the active X and has been shown to be essential for chromosome choice. Today, researchers continue to study the complex Xic region, which is still not fully understood.
The Inactive X Chromosome Has a Unique Chromatin Structure
Sixty years after Barr and Bertram first described the heterochromatic body in the nuclei of female cells, we have learned much about the molecular mechanisms that underlie X inactivation. A model of these events is shown in Figure 7. Binding of XIST RNA, which has been the focus of this article, is thought to be an early event in inactivating the X chromosome. XIST RNA spreads from its site of synthesis at the XIC along the length of the inactive X. More stable inactivation involves the recruitment of the macroH2A histone variant and various silencing complexes, as well as histone modifications common to inactive chromatin (Avner & Heard, 2001). The end point is a chromatin structure that is unique to the inactive X.
References and Recommended Reading
Barr, M. L., & Bertram, E. G. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163, 676–677 (1949) doi:10.1038/163676a0 (link to article)
Brown, C. J., et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44 (1991a) doi:10.1038/349038a0 (link to article)
———. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349, 82–84 (1991b) doi:10.1038/349082a0 (link to article)
Clemson, C. M., et al. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. Journal of Cell Biology 132, 259–275 (1996)
Lee, J. T., & Jaenisch, R. Xist has properties of the X-chromosome inactivation centre. Nature 386, 275–279 (1997) doi:10.1038/386275a0 (link to article)
Lyon, M. F. Gene action in the X chromosome of the mouse. Nature 190, 372–373 (1961) doi:10.1038/190372a0 (link to article)
———. Sex chromatin and gene action in the mammalian X-chromosome. American Journal of Human Genetics 14, 135–148 (1962)
Marahrens, Y., et al. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes and Development 11, 156–166 (1997) doi:10.1101/gad.11.2.156
Martin, G. R., et al. X-chromosome inactivation during differentiation of female teratocarcinoma stem cells in vitro. Nature 271, 329–333 (1978) doi:10.1038/271329a0 (link to article)
Ohno, S., et al. Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Experimental Cell Research 18, 415–418 (1959) doi:10.1016/0014-4827(59)90031-X
Penny, G. D., et al. Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996) doi:10.1038/379131a0 (link to article)
Rastan, S., & Robertson, E. J. X-chromosome deletions in embryo-derived EK lines associated with lack of X-chromosome inactivation. Journal of Embryology and Experimental Morphology 90, 379–388 (1985)



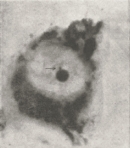
 Figure 1
Figure 1