« Prev Next »
Rearrangements of chromosomes include deletions of DNA sequences and duplications of segments, both of which can encompass thousands to hundreds of thousands of bases. Why do these large rearrangements occur? For one, certain structural features of the genome, also referred to as genome architecture, can render various regions fragile and thus prone to events such as chromosome breakage, which often result in translocations, deletions, and duplications. Often, these alterations happen due to errors during cell division when chromosomes align (Figure 1). Homologous recombination between areas of concentrated repeated sequences frequently creates deletions and duplications. Because they commonly involve more than one gene, the disorders caused by these large deletion and duplication mutations are often severe.
Chromosomal Duplications
In chromosomal duplications, extra copies of a chromosomal region are formed, resulting in different copy numbers of genes within that area of the chromosome. If the duplicated sections are adjacent to the original, the process is known as tandem duplication, whereas if they are separated by nonduplicated regions, the duplication is said to be displaced. Duplications may affect phenotype by altering gene dosage. For example, the amount of protein synthesized is often proportional to the number of gene copies present, so extra genes can lead to excess proteins. Because most embryonic developmental processes are heavily dependent on carefully balanced levels of proteins, duplications resulting in extra gene copies (Figure 1) can therefore lead to developmental defects such as those seen in the Drosophila Bar eye mutation (Figure 2).
Similarly to the effect of other rearrangements, however, duplications can also provide raw material for evolution by producing new copies of genes that are free to mutate and take on other functions. Gene families such as the human globin gene family attest to the role of duplication in evolution. Several globin genes have arisen from a single ancestral precursor, thus making individual genes available to take on specialized roles, with some genes becoming active during embryonic and fetal development, and others becoming active in the adult organism (Figure 3).
Chromosomal Deletions
Deletions involve the loss of DNA sequences. Phenotypic effects of deletions depend on the size and location of deleted sequences on the genome. For instance, deletions that span a centromere result in an acentric chromosome that will most likely be lost during cell division. As with duplications, deletions can affect gene dosage and thus the resulting phenotype. Also, the larger the deletion, the more genes are likely to be involved, and the more drastic the resulting defect is likely to be.
The copy number of specific genes needs to be tightly regulated to ensure that when a gene is expressed, the functional product is produced at the correct level, or dosage. Some genes require two copies for normal expression levels to be produced. If one copy is lacking and only one allele remains, a mutant phenotype results; this is called haploinsufficiency. For example, a group of genetic conditions known as short stature homeobox-related haploinsufficiency disorders results from the loss of one copy of an allele of the SHOX gene. Individuals with these disorders are short in stature and display other physical anomalies (Munns & Glass, 2008).
Clustering of Breakpoints: Recombination Hotspots
Recombination of homologous chromosomes is an important aspect of the generation of genetic variation in species, as well as a normal process that is part of meiosis, specifically occurring between sister chromatids during meiosis I. While allelic homologous recombination (AHR) results in the interchange of homologous genetic material and the separation of haplotypes, there are often cases of nonallelic homologous recombination (NAHR), in which the sequences that recombine are on different chromosomes or different positions of homologous chromosomes. In fact, recombination can even occur between two sequences on the same chromosome. Such instances of NAHR can result in alterations in chromosomal structure, including the generation of deletions and duplications.
Although chromosome rearrangements have been found throughout the human genome, they tend to be most common in specific regions or "hotspots." Often, these hotspots are flanked on either side by areas of low recombination. Historically, recombinatory hotspots were inferentially detected by examining familial pedigrees for regions of the genome that showed recombination more frequently than expected. However, this is difficult to do in human families, in part because they are small and in each individual, recombination occurs on average once every 80 million base pairs. Accurate recombination rates are calculated using thousands of genetic markers that scientists have characterized across an organism's genome. Researchers then divide the genetic distance between markers (measured in centimorgans [cM], or the distance giving rise to 1% recombinant offspring) by the corresponding physical distance (measured in megabases [Mb]). While the recombination rate thus changes over the entire genome, the average rate in humans is approximately 1.25 cM/Mb. More recently, however, scientists have been able to develop higher-throughput methods. Rather than examining families with only a few offspring, they have begun to examine tens of thousands of gametes (sperm cells) at once for evidence of recombination at specific loci (Figure 4; Clark, 2005; Jeffreys et al., 1998).
Scientists are thus discovering that recombination hotspots average about 1.5 kilobases in length and are surrounded by areas of sequence in which recombination appears to be suppressed. Figure 5 schematizes data obtained from a high-throughput study of recombination rates in the human Tap2 hotspot (Jeffreys et al., 2000; Kauppi et al., 2004). Over this area of about 200 kilobases of genomic sequence, there are six hotspots at which recombination occurs much more frequently than the average of once every 1.1 kilobases; however, the rest of the region is "cold," exhibiting little evidence of recombination.

Role of Nonallelic Homologous Recombination in Duplications and Deletions
In some cases, recombination does not result in the exchange of completely homologous sequences, especially in areas that have highly repetitive DNA, like the pericentromeric regions. These regions around the centromere are rich in low-copy repeat (LCR) sequences and are often substrates for NAHR. LCRs are typically 10 to 500 kilobases in length and about 95% identical, consisting of repetitions of the same or closely similar base-pair sequences. Many of them have sequences suggesting that they may have originated in viruses. These sequences have been estimated to constitute 5% to 10% of the genome. In some areas, this percentage is even higher; for instance, a 7.5 million base sequence on the short arm of chromosome 17 is more than 23% LCRs (Stankiewicz et al., 2003). Recombination between inverted LCRs or LCRs on different chromosomes produces forms of genomic rearrangement other than duplications and deletions, such as inversions and translocations (Figure 6).

Deletions, Duplications, and Disease
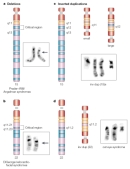
Cytogenetically visible deletions occur in 1 in approximately every 7,000 live births (Jacobs et al., 1992). A number of human disorders are caused by chromosomal deletions, and, generally, their phenotypes are more severe than those caused by duplications (Brewer et al., 1998). Among the better-characterized deletion disorders are cri du chat (French for "cry of the cat") syndrome, named for the distinctive cry affected infants make due in part to malformations of the larynx, and Prader-Willi syndrome (PWS). Both disorders are characterized by mental retardation, as well as a number of physical defects. Cri du chat, which occurs in roughly 1 in every 50,000 live births, results from a deletion on the short arm of chromosome 5 (Chen, 2007). PWS, which occurs at approximately the same frequency, can be caused by a deletion on the long arm of chromosome 15 (Figure 7), although other chromosomal abnormalities can also lead to the syndrome. The genetic basis of PWS is further complicated by the fact that the genetic region involved is also subject to genomic imprinting; that is, the phenotype differs depending upon which parent the deletion is inherited from. Several other diseases related to chromosomal duplications and deletions are listed in Table 1.
Table 1: Examples of Diseases Resulting from Duplications or Deletions of Chromosomal Regions
| Genetic Disease | Type of Rearrangement | Location Affected |
| Charcot-Marie-Tooth disease type I | Duplication | 17p12 |
| Hereditary neuropathy with pressure palsies | Deletion | 17p12 |
| Smith-Magenis syndrome | Deletion | 17p11.2 |
| Williams-Beuren syndrome | Deletion | 7q11.23 |
Because deletions and duplications are often central to our understanding of many human disorders, knowledge of the mechanisms behind these genetic changes is a high priority. The Human Genome Project has shown that changes in the numbers of copies of many sequences are much more common than previously imagined. All of these factors suggest that the study of duplications and deletions is ripe for major advancement.
References and Recommended Reading
Amos-Landgraf, J. M., et al. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. American Journal of Human Genetics 65, 370–386 (1999)
Bailey, J. A., et al. Primate segmental duplications: Crucibles of evolution, diversity, and disease. Nature Reviews Genetics 7, 552–564 (2006) doi:10.1038/nrg1895 (link to article)
Brewer, C., et al. A chromosomal deletion map of human malformation. American Journal of Human Genetics 63, 1153–1159 (1998)
Chance, P. F., et al. Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Human Molecular Genetics 3, 223–228 (1994)
Chen, H. Cri du chat syndrome. eMedicine (2007) (link to article)
Chen, K. S., et al. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nature Genetics 17, 154-163 (1997) doi:10.1038/ng1097-154 (link to article)
Christian, S. L., et al. Large genomic duplicons map to sites of instability in Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Human Molecular Genetics 8, 1025–1037 (1999)
Clark, A. G. Hot spots unglued. Nature Genetics 37, 563–564 (2005) doi:10.1038/ng0605-563 (link to article)
Emanuel, B. S., & Shaikh, T. H. Segmental duplications: An "expanding" role in genomic instability and disease. Nature Reviews Genetics 2, 791–800 (2001) doi:10.1038/35093500 (link to article)
Hedrick, P. W. Inference of recombination hotspots using gametic disequilibrium values. Heredity 60, 435–438 (1988)
Jacobs, P. A., et al. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. Journal of Medical Genetics 29, 103–108 (1992)
Jeffreys, A. J., et al. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Molecular Cell 2, 267–273 (1998)
Jeffreys, A. J., et al. High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Human Molecular Genetics 9, 725–733 (2000)
Kauppi, A., et al. Where the crossovers are: Recombination distributions in mammals. Nature Reviews Genetics 5, 413–424 (2004) doi:10.1038/nrg1346 (link to article)
Kong, A. et al. Recombination rate and reproductive success in humans. Nature Genetics 36, 1203–1206 (2004) doi:10.1038/ng1445 (link to article)
Lupski, J. R. Charcot-Marie-Tooth disease: Lessons in genetic mechanisms. Molecular Medicine 4, 3–11 (1998)
Lupski, J. R., & Stankiewicz, P. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genetics 1, 627–633 (2005)
Munns, C., & Glass, I. SHOX-related haploinsufficiency disorders. Gene Reviews (2008)
Perez-Jurado, L. A., et al. A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Human Molecular Genetics 7, 325–334 (1998)
Peoples, R., et al. A physical map, including a BAC/PAC clone contig, of the Williams-Beuren syndrome-deletion region at 7q11.23. American Journal of Human Genetics 66, 47–68 (2000)
Ptak, S. E., et al. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biology 2, 849–855 (2004)
Shaw, C. J., & Lupski, J. R. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Human Molecular Genetics 13, R57–R64 (2004)
Stankiewicz, P., et al. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. American Journal of Human Genetics 72, 1101–1116 (2003)




 Figure 2
Figure 2