« Prev Next »
Most cytogeneticists are skilled at identifying individual chromosomes based on their sizes, their shapes, and the banding patterns of their arms. Still, the ability to pinpoint particular chromosomes or regions thereof is of little use unless researchers also have a uniform system for referring to these structures in their communications with one another. This is precisely where the practice of chromosome mapping comes into play.
Today, researchers all over the world rely on a regularly updated mapping system that allows them to refer to specific portions of particular chromosomes with extreme accuracy. This system follows international standards, and it revolves around use of the schematic diagram known as an idiogram, or ideogram. (Both spellings of the word are commonly found in scientific literature.) Idiograms provide a pictorial reference point that is useful for locating the positions of individual genes on chromosomes, as well as for identifying various abnormalities associated with a range of chromosomal disorders. Moreover, idiograms enable members of the scientific community to reference important sources, like the Human Genome Project, through a universal vocabulary that allows for quick, clear interpretation.
Understanding the ISCN Mapping System
A consistent numbering system for mapping genes on chromosomes is essential for producing relevant idiograms. Researchers rely on a mapping system known as the International System for Cytogenetic Nomenclature (ISCN) for this purpose. In the ISCN scheme, the numbering system for a chromosome begins at its centromere. Chromosomes are assigned a long arm and a short arm, based on the position of their centromeres. The shorter arm of the chromosome is known as the p, or petite arm, from the French word for "small." The longer arm is known as the q, or queue arm, from the word meaning a line of people. Thus, chromosomal regions that are present on the short arm will begin with the designation p, whereas regions on the long arm will begin with q. By convention, the p arm of the chromosome is always shown at the top in a karyotype.
Each arm of the chromosome is then divided into regions, and the numbers assigned to each region get larger as the distance from the centromere to the telomere increases. Regions are identified by the specific morphological features that are consistently found in a chromosome, such as the presence of prominent Giemsa-staining bands. The regions are named p1, p2, etc., on the short arm and q1, q2, etc., on the long arm. Depending on the resolution of the staining procedure, it may be possible to detect additional bands within each region, which are designated by adding another digit to the number of the region, once again increasing in value as the distance from the centromere increases.
The Mapping System in Practice
A specific example helps illustrate how the chromosome numbering system works. Figure 1 shows an idiogram for chromosome 12, a medium-sized chromosome with one long and one short arm. The position of the centromere, which separates the p and q arms, is shown by the hatched area. This particular idiogram depicts the pattern of Giemsa staining at a fairly low resolution (i.e., it produces about 400 total bands in a karyotype, which is just above the threshold that is clinically useful). At this resolution, the long q arm of chromosome 12 can be subdivided into two main regions, which are designated 12q1 and 12q2. Region 12q1 can be further subdivided into five subregions, designated 12q11 through 12q15, each of which corresponds to a band detected by Giemsa staining. Orally, these subdivisions are referred to as "12q one-one" through "12q one-five" (not as "12q eleven" through "12q fifteen"). The more distal 12q2 region can be subdivided into subregions 12q21 through 12q24. In addition, subregion 12q24 can be further subdivided into regions 12q24.1 through 12q24.3, even at this relatively low resolution.
Higher resolution views of the chromosome can be obtained if staining is done while the chromosomes are in prometaphase and are less condensed. At this higher level of resolution, approximately 850 bands can be distinguished in a karyotype. As Figure 2 illustrates, additional subdivisions can be detected in all of the regions on chromosome 12 under these conditions.
The Human Genome Project Provides the Ultimate Chromosome Map
Once a defect has been associated with a particular chromosomal position by traditional cytogenetics, investigators are often interested in identifying candidate genes within that region that may be responsible for the symptoms displayed by a patient. With the completion of the Human Genome Project (HGP), this process has been greatly accelerated. The HGP data provide a map of all human chromosomes at single-nucleotide resolution, and the positions of individual genes have been correlated with specific bands detected on high-resolution idiograms of individual chromosomes. Indeed, the Graphic Chromosome Map Viewer available from the National Center for Biotechnology Information provides a simple link between the cytogenetic and sequence data.
Summary
As scientists work to correlate specific genes with specific chromosomal bands, idiograms are becoming increasingly important. Idiograms ensure consistent identification of regions within chromosomes, thereby enabling consistent dialogue and easy cross-referencing between individual samples and reference sources. They also provide links between structural abnormalities and the individual genes correlated with various diseases or syndromes. Thus, idiograms are essential to our ability to organize genomic information as we move into an increasingly molecular era of cytogenetics.
References and Recommended Reading
Braude, P., et al. Preimplantation genetic diagnosis. Nature Reviews Genetics 3, 941–953 (2002) doi:10.1038/nrg953 (link to article)
Scherer, S. E., et al. The finished DNA sequence of human chromosome 12. Nature 440, 346–351 (2006) doi:10.1038/nature04569 (link to article)
Strachan, T., & Read, A. P. Human Molecular Genetics, 2nd ed. (New York, Wiley, 1999)



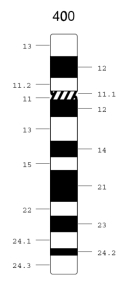
 Figure 1
Figure 1