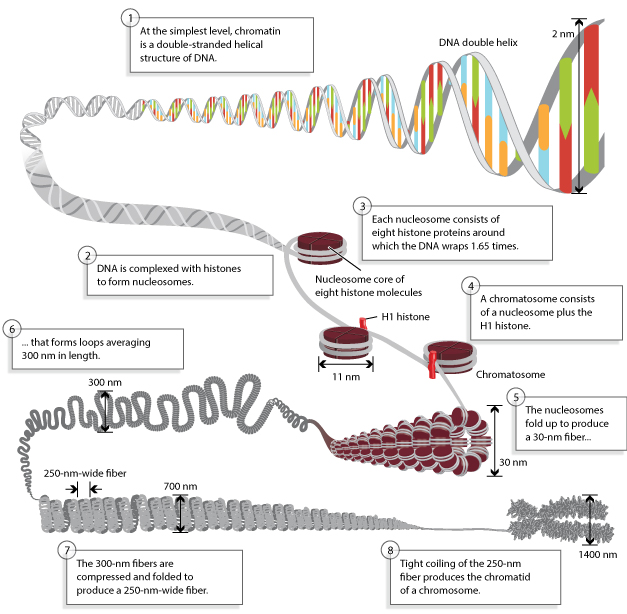
« Prev Next »
The haploid human genome contains approximately 3 billion base pairs of DNA packaged into 23 chromosomes. Of course, most cells in the body (except for female ova and male sperm) are diploid, with 23 pairs of chromosomes. That makes a total of 6 billion base pairs of DNA per cell. Because each base pair is around 0.34 nanometers long (a nanometer is one-billionth of a meter), each diploid cell therefore contains about 2 meters of DNA [(0.34 × 10-9) × (6 × 109)]. Moreover, it is estimated that the human body contains about 50 trillion cells—which works out to 100 trillion meters of DNA per human. Now, consider the fact that the Sun is 150 billion meters from Earth. This means that each of us has enough DNA to go from here to the Sun and back more than 300 times, or around Earth's equator 2.5 million times! How is this possible?
DNA, Histones, and Chromatin
Histones are a family of small, positively charged proteins termed H1, H2A, H2B, H3, and H4 (Van Holde, 1988). DNA is negatively charged, due to the phosphate groups in its phosphate-sugar backbone, so histones bind with DNA very tightly.
The Nucleosome: The Unit of Chromatin
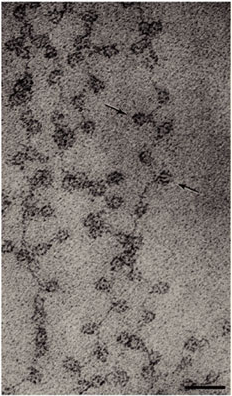
Today, researchers know that nucleosomes are structured as follows: Two each of the histones H2A, H2B, H3, and H4 come together to form a histone octamer, which binds and wraps approximately 1.7 turns of DNA, or about 146 base pairs. The addition of one H1 protein wraps another 20 base pairs, resulting in two full turns around the octamer, and forming a structure called a chromatosome (Box 4 in Figure 1). The resulting 166 base pairs is not very long, considering that each chromosome contains over 100 million base pairs of DNA on average. Therefore, every chromosome contains hundreds of thousands of nucleosomes, and these nucleosomes are joined by the DNA that runs between them (an average of about 20 base pairs). This joining DNA is referred to as linker DNA. Each chromosome is thus a long chain of nucleosomes, which gives the appearance of a string of beads when viewed using an electron microscope (Figure 2; Olins & Olins, 1974, 2003).
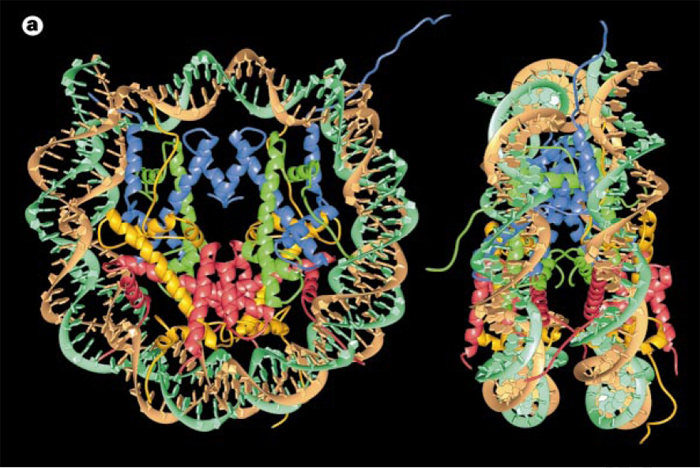
The amount of DNA per nucleosome was determined by treating chromatin with an enzyme that cuts DNA (such enzymes are called DNases). One such enzyme, micrococcal nuclease (MNase), has the important property of preferentially cutting the linker DNA between nucleosomes well before it cuts the DNA that is wrapped around octamers. By regulating the amount of cutting that occurs after application of MNase, it is possible to stop the reaction before every linker DNA has been cleaved. At this point, the treated chromatin will consist of mononucleosomes, dinucleosomes (connected by linker DNA), trinucleosomes, and so forth (Hewish and Burgoyne, 1973).If DNA from MNase-treated chromatin is then separated on a gel, a number of bands will appear, each having a length that is a multiple of mononucleosomal DNA (Noll, 1974). The simplest explanation for this observation is that chromatin possesses a fundamental repeating structure. When this was considered together with data from electron microscopy and chemical cross-linking of histones, the "subunit theory" of chromatin (Kornberg, 1974; Van Holde et al., 1974) was adopted. The subunits were later named nucleosomes (Oudet et al., 1975) and were eventually crystallized (Luger et al., 1997). The model of the nucleosome that crystallographers constructed from their data is shown in Figure 3. Phosphodiester backbones of the DNA double helix are shown in brown and turquoise, while histone proteins are shown in blue (H3), green (H4), yellow (H2A), and red (H2B). Note that only eukaryotes (i.e., organisms with a nucleus and nuclear envelope) have nucleosomes. Prokaryotes, such as bacteria, do not.
Chromatin Is Coiled into Higher-Order Structures
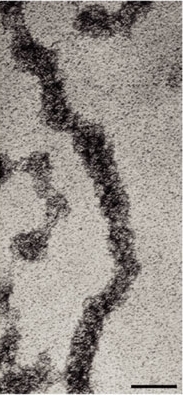
Over the years, there has been a great deal of speculation concerning the manner in which nucleosomes are folded into 30-nanometer fibers (Woodcock, 2005). Part of the problem lies in the fact that electron microscopy is perhaps the best way to visualize packaging, but individual nucleosomes are hard to discern after the fiber has formed. In addition, it also makes a difference whether observations are made using isolated chromatin fibers or chromatin within whole nuclei. Thus, the 30-nanometer fiber may be highly irregular and not quite the uniform structure depicted in instructive drawings such as Figure 1 (Bednar et al., 1998). Interestingly, histone H1 is very important in stabilizing chromatin higher-order structures, and 30-nanometer fibers form most readily when H1 is present.
Processes such as transcription and replication require the two strands of DNA to come apart temporarily, thus allowing polymerases access to the DNA template. However, the presence of nucleosomes and the folding of chromatin into 30-nanometer fibers pose barriers to the enzymes that unwind and copy DNA. It is therefore important for cells to have means of opening up chromatin fibers and/or removing histones transiently to permit transcription and replication to proceed. Generally speaking, there are two major mechanisms by which chromatin is made more accessible:
- Histones can be enzymatically modified by the addition of acetyl, methyl, or phosphate groups (Fischle et al., 2005).
- Histones can be displaced by chromatin remodeling complexes, thereby exposing underlying DNA sequences to polymerases and other enzymes (Smith & Peterson, 2005).
It is important to remember that these processes are reversible, so modified or remodeled chromatin can be returned to its compact state after transcription and/or replication are complete.
Chromosomes Are Most Compacted During Metaphase
When eukaryotic cells divide, genomic DNA must be equally partitioned into both daughter cells. To accomplish this, the DNA becomes highly compacted into the classic metaphase chromosomes that can be seen with a light microscope. Once a cell has divided, its chromosomes uncoil again.
Comparing the length of metaphase chromosomes to that of naked DNA, the packing ratio of DNA in metaphase chromosomes is approximately 10,000:1 (depending on the chromosome). This can be thought of as akin to taking a rope as long as a football field and compacting it down to less than half an inch. This level of compaction is achieved by repeatedly folding chromatin fibers into a hierarchy of multiple loops and coils (Figure 1). Exactly how this is accomplished is unclear, but the phosphorylation of histone H1 may play a role. Indeed, this is just one area of DNA packaging that researchers will continue to explore in the years to come.
References and Recommended Reading
Bednar, J., et al. Nucleosomes, linker DNA, and linker histones form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proceedings of the National Academy of Sciences 95, 14173–14178 (1998)
Fischle, W., et al. Histone and chromatin cross-talk. Current Opinion in Cellular Biology 15, 172–183 (2003)
Hewish, D. R., and Burgoyne, L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun 52, 504-510 (1973)
Kornberg, R. D. Chromatin structure: A repeating unit of histones and DNA. Science 184, 868–871 (1974)
Luger, K., et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997) (link to article)
Noll, M. Subunit structure of chromatin. Nature 251, 249–251 (1974) (link to article)
Olins, A. L., & Olins, D. E. Spheroid chromatin units (v bodies). Science 183, 330–332 (1974)
Olins, D. E., & Olins, A. L. Chromatin history: Our view from the bridge. Nature Reviews Molecular Cell Biology 4, 809–814 (2003) (link to article)
Oudet, P., et al. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4, 281–300 (1975)
Smith, C. L., & Peterson, C. L. ATP-dependent chromatin remodeling. Current Topics in Developmental Biology 65, 115–148 (2005)
Thomas, J. O., & Kornberg, R. D. Octamer of histones in chromatin and free in solution. Proceedings of the National Academy of Sciences 72, 2626–2630 (1975)
Van Holde, K. E. Chromatin: Springer Series in Molecular Biology (New York, Springer-Verlag, 1988)
Van Holde, K. E., et al. A model for particulate structure in chromatin. Nucleic Acids Research 1, 1579–1586 (1974)
Wolffe, A. P. Chromatin: Structure and Function, 3rd ed. (San Diego, Academic, 1999)
Woodcock, C. L. A milestone in the odyssey of higher-order chromatin structure. Nature Structural and Molecular Biology 12, 639–640 (2005) (link to article)
Woodcock, C. L., et al. Structural repeating units in chromatin. I. Evidence for their general occurrence. Experimental Cell Research 97, 101–110 (1976)