« Prev Next »
Chances are that when most people hear the word "chromosome," they picture a rodlike, highly compacted chunk of DNA. While chromosomes do appear as condensed, elongated structures during the process of cell division, for most of a cell's lifetime, chromosomes do not look anything like this (Figure 1). In fact, some cell types, such as neurons, never undergo cell division, and their chromosomes therefore never take on a condensed state. So, what do chromosomes look like in the nucleus of a cell between cell divisions? Although this seems to be a simple question, its answer has only been revealed within the last two decades, and the implications of what has been found are both fascinating and the subject of much ongoing work in the field of genome cell biology.
A Technically Challenging Problem: Determining How Chromosomes Are Organized
Determining how chromosomes are organized inside the cellular nucleus was a technically challenging problem for scientists for many decades. One might think that it would be possible to simply use microscopes to peer into the nucleus and see how chromosomes are arranged. Indeed, during the 1960s and 1970s, various approaches involving both electron and light microscopy showed that chromosomes undergo a dramatic structural change as cells transition from mitosis to interphase, and that these structures exist as decondensed structures between cell divisions. However, microscopy did not reveal how chromosomes were organized in the nucleus of cells that were not undergoing mitosis. In fact, during the period between cell divisions, no distinct individual chromosomes can be recognized via either light or electron microscopy; thus, it is impossible to distinguish one chromosome from its neighbor using these methods.
Models of Chromosome Organization
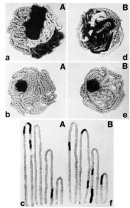
Despite a lack of direct observational evidence, early researchers proposed two models for the way in which chromosomes were likely organized in the nuclei of nondividing cells. The first model, known as the chromosome territory model, was originally proposed by Carl Rabl in 1885. According to this model, the DNA of each chromosome occupies a defined volume of the nucleus and only overlaps with its immediate neighbors (Figure 2Ab). In contrast, according to the second or "spaghetti" model, the DNA fiber of multiple chromosomes meanders through the nucleus in a largely random fashion, and the chromosomes are therefore intermingled and entangled with each other (Figure 2Be).
Testing the Models
The key experiment to distinguish between these two models was eventually carried out in the early 1980s by Thomas Cremer, a German cell biologist, and his physicist brother, Christoph Cremer. The Cremers realized that the two models made very distinct predictions and could thus be distinguished from each other if it were somehow possible to mark chromosome regions in part of the nucleus and follow the fate of these regions during cell division. Specifically, the Cremers argued that if chromosomes existed in territories occupying only a limited volume in the nucleus, marking of a defined volume would only affect very few immediately adjacent chromosomes (Figure 2Aa). On the other hand, if the spaghetti model were correct, the marking of a small volume would affect a large number of chromosomes, because fibers from many different chromosomes would be running through the marked volume (Figure 2Bd).
To carry out this decisive experiment, Christoph Cremer and his group developed a laser that could be focused very narrowly to shine on a small section of a cell's nucleus. The laser light was of a wavelength that induced DNA damage in the illuminated regions. The Cremers then exploited the ability of cells to repair damaged DNA by providing radioactively labeled nucleotides, which the cell incorporated into its DNA during the repair process. Then, when the cell entered the next mitosis and its chromosomes took on a condensed appearance, the marked regions were analyzed by radiography.
The results were clear. The Cremer brothers found that only a few chromosomes per cell were damaged, a result that strongly supported the chromosome territory model. The existence of chromosome territories was impressively confirmed a few years later by the development of the fluorescence in situ hybridization (FISH) technique, in which fluorescently labeled probes complementary to a specific chromosome are used to visualize a given chromosome in the intact nucleus. These experiments allowed direct visualization of the territorial nature of chromosomes (Figure 3).
What FISH Reveals
Since its inception, the FISH technique has become one of the workhorse methods in the field of genome cell biology, and use of this technique has revealed many fundamental properties of chromosome territories. For instance, in combination with high-resolution microscopy, FISH has demonstrated that chromosome territories are irregular in shape but typically about 1 to 2 micrometers in diameter, and they consist of smaller subdomains. We now also know that chromosome territories exist in all higher eukaryotes, although chromosomes are more unfolded in lower eukaryotes such as yeast. Chromosome territories have additionally been found to border each other closely; in fact, neighboring chromosomes can invade each other's territories and intermingle at their peripheries. Observations of living cells have also revealed that chromosomes are essentially immobile, most likely held in place by the force exerted upon them by their neighbors. Furthermore, chromosome territories are semiconserved from parent to daughter cell during cell division, with locations in the daughter cell similar to those in the parent cell (Parada et al., 2003).
The Arrangement and Alteration of Chromosome Territories
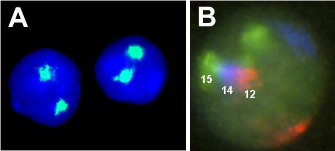
The fact that each chromosome occupies a distinct region of a cell's nucleus raises a fundamental question: Are chromosome territories randomly arranged in the nucleus, or they are they organized into patterns? Two experiments demonstrate that chromosomes are not randomly localized within the nucleus. First, when researchers measure the distance of a given chromosome from the center of a cell's nucleus, they note that some chromosomes localize toward the periphery, often touching the nuclear membrane, whereas others are located toward the center of the nucleus. Second, when visualizing multiple chromosome territories, scientists tend to see recurrent clusters of chromosomes. For example, in mouse lymphocytes, chromosome 12 often sits next to chromosome 14, which in turn is adjacent to chromosome 15, thereby forming a triplet cluster (Figure 3B).
Researchers have also examined whether various findings related to chromosomal territories hold true across species. Through such studies, scientists have noted that there are large areas of chromosomal identity between different species that have been maintained throughout evolution; moreover, these areas of identity maintain their positions in different species (Tanabe et al., 2002). Indeed, analysis of chromosome territories in many cell types and tissues has made it clear that patterns of chromosome arrangement are specific to both cell type and tissue type.
Yet another interesting observation has been the finding that chromosome territories can reposition in disease, which might provide novel insights into disease mechanisms and why genes are incorrectly expressed in disease. In fact, scientists have manipulated the localization of chromosomes and seen some changes in gene expression as a result, thus suggesting a possible mechanism for the connection between chromosomal territories and disease (Finlan et al., 2008).
Despite these discoveries, researchers still have numerous questions about chromosome territories. The holy grail of the field, for instance, is to understand what mechanisms determine exactly where a chromosome ends up in the nucleus. As of July 2008, no proteins have been identified that either anchor chromosomes in the nucleus or link multiple chromosomes to each other to establish chromosome clusters. As research continues, perhaps these and other questions will eventually be answered.
References and Recommended Reading
Cremer, T., & Cremer, C., Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Reviews Genetics 2, 292–301 (2001) doi:10.1038/35066075 (link to article)
Cremer, T., et al. Rabl's model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Human Genetics 60, 46–56 (1982)
Finlan, L. E., et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genetics 4, e1000039 (2008) doi:10.1371/journal.pgen.1000039 (link to article)
Lanctôt, C., et al. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nature Reviews Genetics 8, 104–115 (2007) doi:10.1038/nrg2041 (link to article)
Meaburn, K. J., & Misteli, T. Cell Biology: Chromosome territories. Nature 445, 379–381 (2007) doi:10.1038/445379a (link to article)
Parada, L. A., et al. Conservation of relative chromosome positioning in normal and cancer cells. Current Biology 12, 1692–1697 (2002)
———. An uncertainty principle in chromosome positioning. Trends in Cell Biology 13, 393–396 (2003)
———. Tissue-specific spatial organization of genomes. Genome Biology 5, R44 (2004)
Tanabe, H., et al. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proceedings of the National Academy of Sciences 99, 4424–4429 (2002)



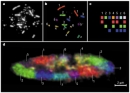
 Figure 1
Figure 1