« Prev Next »
Telomeres Protect Chromosomes from End-to-End Fusions
McClintock analyzed the consequences of chromosome breakage by constructing a maize strain in which dicentric chromosomes (those containing two centromeres) could be produced with high frequency. Dicentric chromosomes break when the two centromeres are drawn to opposite poles of the mitotic spindle during cell division. In her work with this maize strain, McClintock noted that the broken chromosomal ends were unstable, and that these ends fused with any other broken ends with which they came in contact. If the original chromosome reformed when such contact occurred, then the cycle of breakage and fusion could be repeated. This cycle was broken, however, when dicentric chromosomes were present in embryonic cells; in these cells, the broken ends were somehow "healed" (McClintock, 1941). Today, researchers know that the ends were healed by the addition of a telomere, a process that is catalyzed by an enzyme called telomerase. Telomerase is active in germ cells, embryonic cells, and some somatic cells, but not in the endosperm cells that McClintock examined (McKnight & Shippen, 2004).
Thus, in her experiments, McClintock observed several fundamental properties of chromosome biology; specifically, she noticed that cells do not tolerate the presence of unprotected chromosome ends, and that broken ends are quickly fused together by the DNA repair machinery. McClintock also saw that telomeres prevent these kinds of fusion events from occurring. Despite the magnitude of these discoveries, 40 years would elapse between McClintock's original observations and the experiments that revealed the actual structure of telomeres.
Telomeres Have Many Tandem Repeats of Short GT-Rich Sequences
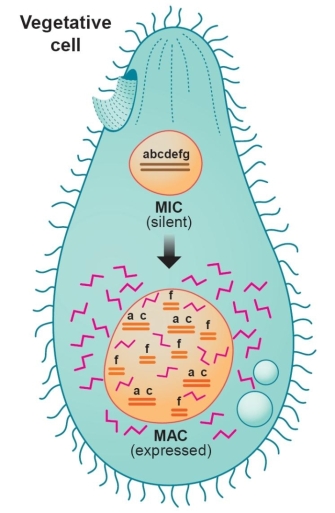
Nature provided the model that gave scientists their first glance into the unique structure of the telomere. Tetrahymena is a large unicellular protozoan with two nuclei, the macronucleus and the micronucleus (Figure 1). The micronucleus is the germ-line nucleus that contains five normal pairs of chromosomes, but it is transcriptionally inactive under most circumstances. The larger macronucleus, which is responsible for programming most of the cell's activities, is derived from a copy of the micronucleus produced by a complex process that selectively fragments the germ-line DNA into 200-300 pieces, ranging in size from a few hundred to a few thousand base pairs. These "minichromosomes," which have telomeres at either end but no centromere, are then amplified many times as the macronucleus matures. Consequently, each macronucleus is estimated to contain about 10,000 minichromosomes.
In the mid-1970s, Elizabeth Blackburn, who was working as a postdoctoral associate in Joseph Gall's laboratory, set out to determine the telomere sequences of a minichromosome that contained the Tetrahymena ribosomal DNA sequence. Approximately 200 copies of this particular minichromosome are present in each cell, and the Gall lab had already worked out the conditions necessary for physically separating this minichromosome from the other minichromsomes in the cell. When Blackburn carried out sequencing experiments for the ribosomal DNA minichromosome, she was surprised to find that the telomeres of this structure contained 20-70 tandem copies of a simple hexanucleotide with the sequence 5'-CCCCAA-3' on one strand and 5'-TTGGGG on the complementary strand. This sequence can also be written CCCCAA/TTGGGG. Significantly, the GT-rich strand represented the 3'-end of the minichromosome (Blackburn & Gall, 1978).
Researchers now know that tandem repeats of short GT-rich sequences are characteristic of almost all eukaryotic telomeres (Vega et al., 2003; de Lange, 2004; McKnight & Shippen, 2004). In general, telomeres consist of a 6-8 base-pair sequence that is repeated hundreds or thousands of times. The actual repeated sequence and the number of repeats vary between species. Human telomeres, for example, range in size from 2-50 kilobases and consist of approximately 300-8,000 precise repeats of the sequence CCCTAA/TTAGGG. By contrast, telomeres from the budding yeast Saccharomyces cerevisiae are smaller and more heterogeneous in their composition. S. cerevisiae telomeres typically contain about 60-100 copies of the sequence C1-3A/TG1-3. A common feature of all telomeres, however, is the orientation of the G-rich strand. In all cases, this strand makes up the 3'-end of the chromosome, and the terminal portion of the G-rich strand is single-stranded, generating a so-called "G-tail." The actual length of the G-tail is somewhat variable, averaging between 75-300 nucleotides in humans and 50-100 nucleotides in yeast.
The Loop at the End of the Chromosome
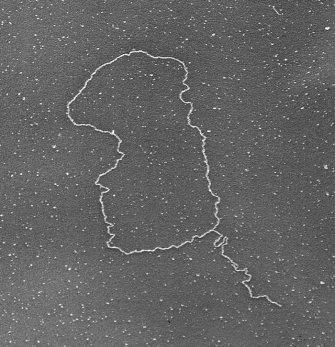
Not surprisingly, the model in Figure 3 does not apply to all eukaryotes with GT-rich telomeres. Although t-loops are highly conserved, their actual structure varies between species. For instance, in some species, notably S. cerevisiae, the G-tail does not invade double-stranded DNA sequences in the telomere. Instead, the t-loop is stabilized primarily by protein-protein interactions, and the G-tail is protected by a single-stranded DNA binding protein. Moreover, Tetrahymena, which has been an important model for telomere biology, presents an interesting paradox. The chromosomes in the Tetrahymena micronucleus terminate in t-loops, but the minichromosomes in its macronucleus lack t-loops and are simply capped by a tenacious binding protein. One could argue that the minichromosomes are not true chromosomes, because they lack centromeres and are not passed down to progeny. Nonetheless, the common theme throughout all species is that telomeres are unique complexes of DNA and proteins that protect the ends of chromosomes.

DNA Replication and Telomerase: End-Replication Problem 'Solved'
When the mechanism of cellular DNA replication was clarified in the early 1970s, scientists realized that this mechanism presented a fundamental problem—specifically, the ends of chromosomes should progressively shorten with each round of DNA replication. This so-called "end-replication" problem, which is graphically depicted in Figure 4a, is a direct consequence of DNA polymerase's biochemical properties. DNA polymerase requires short RNA primers to initiate replication, and it then extends the primers in a 5'-to-3'-direction. Thus, as the replication fork moves along the chromosome, one of the two daughter strands is synthesized continuously. The other daughter strand, known as the lagging strand, is synthesized discontinuously in short fragments known as Okazaki fragments, each of which has its own RNA primer. The RNA primers are subsequently degraded, and the gaps between the Okazaki fragments are then filled in by the DNA repair machinery. A problem arises at the end of the chromosome, however, because the DNA repair machinery is unable to repair the gap left by the terminal RNA primer. Consequently, the new DNA molecule is shorter than the parent DNA molecule by at least the length of one RNA primer. Without a solution to this end-replication problem, chromosomes would progressively shorten over many cell divisions, a process that would bring about catastrophic consequences.
By the mid-1980s, scientists had begun to accumulate evidence that cells were able to solve this end-replication problem by lengthening their telomeres. In a collaborative set of experiments, the Blackburn and Szostak labs discovered that sequences from Tetrahymena could function as telomeres for linear plasmids that were introduced into yeast cells (Blackburn et al., 2006). Furthermore, these researchers discovered that yeast cells elongated the Tetrahymena telomere sequences. The teams received an even bigger surprise when they determined the sequence of the new telomeres. Quite unexpectedly, the elongated telomeres had repeated copies of the yeast TG1-3 repeat, rather than the Tetrahymena TTGGGG repeat. But how were these cells able to elongate telomere sequences from another organism with copies of that organism's own telomere repeat?
The discovery of telomerase provided the answer to this question. Carol Greider, then a graduate student in Blackburn's lab, decided to look for an activity in Tetrahymena extracts that could add nucleotides to a synthetic oligonucleotide that contained four copies of the Tetrahymena telomere repeat. Because of the close resemblance of this oligonucleotide to an actual telomere, Greider was able to purify an enzyme that could lengthen telomeres (Greider & Blackburn, 1985). This enzyme, which was later named telomerase, turns out to be a highly specialized reverse transcriptase, or an enzyme that synthesizes DNA from an RNA template. Telomerase can also be categorized as a ribonucleoprotein, because the RNA template is an integral part of the telomerase complex itself. This RNA template includes a sequence that is complementary to the telomere repeat unit in the same organism, which explains why yeast cells add yeast repeats to a Tetrahymena telomere.
Figure 4b demonstrates how telomerase works. Telomerase binds to the G-tail of the telomere through the RNA template, and it then catalyzes the extension of the G-tail. Only one extension cycle is illustrated in this figure, but in reality, telomerase is able to repeat this cycle multiple times by moving to new binding sites along the newly synthesized G-tail. Then, during the next round of DNA replication, DNA polymerase and the DNA repair enzymes fill in the other strand. Thus, telomerase is able to both maintain and extend the length of telomeres.
Since the discovery of telomerase, it has become clear that this substance plays a key role in the regulation of telomere length. In general, telomeres tend to shorten in cells without telomerase over time, and cells may stop dividing and become senescent after their telomeres shorten below a critical length. A similar result is seen in knockout mice that lack telomerase activity (Blasco et al., 1997; Lee et al., 1998). Progressive telomere shortening is observed in each generation of these mice, although tissues do not show visible defects until the sixth generation, when the animals' telomeres are very short or undetectable. Some of the first abnormalities appear in reproductive and hematopoietic tissues, which is consistent with the normal tissue distribution of telomerase in mice. Indeed, in most animals (including mice), embryonic cells and germ cells possess telomerase activity, but many types of somatic cells do not. The exceptions include highly proliferative cells, such as those in the skin, hematopoietic tissues, and the intestinal epithelium.
At the cellular level, telomerase-deficient mice show chromosomal abnormalities several generations before defects become apparent at the tissue level. For example, when cells from fourth-generation animals were cultured, investigators noted increased levels of chromosomal abnormalities compared to controls. Interestingly, some of the cells from knockout mice showed end-to-end fusions, confirming Muller and McClintock's early predictions about telomere function.
Telomere Biology Is an Active Area of Research
Our view of the telomere has matured considerably since Blackburn and Gall provided the first information about its molecular composition. We now appreciate that this unusual complex of DNA and protein is a dynamic structure that depends on telomerase and other cellular factors for maintenance. Regulation of telomere function is an active area of current research, and new insights into telomeres and their role in aging and cancer appear regularly in the scientific literature. One thing is clear, however: We haven't yet heard the end of the telomere story.
References and Recommended Reading
Blackburn, E. H., & Gall, J.C. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. Journal of Molecular Biology 120, 33–53 (1978) doi:10.1016/0022-2836(78)90294-2
Blackburn, E. H., et al. Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nature Medicine 12, 1133–1138 (2006) doi:10.1038/nm1006-1133 (link to article)
Blasco, M.A., et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34 (1997) doi:10.1016/S0092-8674(01)80006-4
de Lange, T. T-loops and the origin of telomeres. Nature Reviews Molecular Cell Biology 5, 323–329 (2004) doi:10.1038/nrm1359 link to article)
Greider, C. W., & Blackburn, E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413 (1985)
Griffith, J. D., et al. Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 (1999) doi:10.1016/S0092-8674(00)80760-6
Lee, H. W., et al. Essential role of mouse telomerase in highly proliferative organs. Nature 392, 569–574 (1998) doi:10.1038/33345 (link to article)
McClintock, B. The stability of broken ends of chromosomes in Zea mays. Genetics 26, 234–282 (1941)
McKnight, T. D., & Shippen, D. E. Plant telomere biology. Plant Cell 16, 794–803 (2004)
Neumann, A. A., & Reddel, R. R. Telomere maintenance and cancer—Look, no telomerase. Nature Reviews Cancer 2, 879–884 (2002) doi:10.1038/nrc929 (link to article)
Selker, E. U. A self-help guide for a trim genome. Science 300, 1517–1518 (2003) doi:10.1126/science.1086053
Vega, L. R., et al. Getting to the end: Telomerase access in yeast and humans. Nature Reviews Molecular Cell Biology 4, 948–959 (2003) doi:10.1038/nrm1256 (link to article).



 Figure 2: T-loop.
Figure 2: T-loop.