Abstract
The parasite Plasmodium knowlesi has been the sole cause of malaria in Malaysia from 2018 to 2022. The persistence of this zoonotic species has hampered Malaysia’s progress towards achieving the malaria-free status awarded by the World Health Organisation (WHO). Due to the zoonotic nature of P. knowlesi infections, it is important to study the prevalence of the parasite in the macaque host, the long-tailed macaque (Macaca fascicularis). Apart from P. knowlesi, the long-tailed macaque is also able to harbour Plasmodium cynomolgi, Plasmodium inui, Plasmodium caotneyi and Plasmodium fieldi. Here we report the prevalence of the 5 simian malaria parasites in the wild long-tailed macaque population in 12 out of the 13 states in Peninsular Malaysia using a nested PCR approach targeting the 18s ribosomal RNA (18s rRNA) gene. It was found that all five Plasmodium species were widely distributed throughout Peninsular Malaysia except for states with major cities such as Kuala Lumpur and Putrajaya. Of note, Pahang reported a malaria prevalence of 100% in the long-tailed macaque population, identifying it as a potential hotspot for zoonotic transmission. Overall, this study shows the distribution of the 5 simian malaria parasite species throughout Peninsular Malaysia, the data of which could be used to guide future malaria control interventions to target zoonotic malaria.
Similar content being viewed by others
Introduction
Malaria is still a significant health burden in many countries around the world with 249 million cases reported globally and 608,000 deaths in 20221. This disease is transmitted by Anopheles mosquitoes and is primarily caused by six Plasmodium species, which are Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale curtisi, Plasmodium ovale wallikeri and Plasmodium knowlesi. In Malaysia, from the years 2018 to 2022, P. knowlesi has been the sole cause of all indigenous malaria cases. The persistence of P. knowlesi cases in Malaysia, accounting for 2505 cases and 9 deaths in 2022 alone, has prevented Malaysia from achieving the malaria-free status1 by the World Health Organisation.
Plasmodium knowlesi is considered a zoonotic malaria species as the parasite originally resides in a macaque host. The currently known hosts of P. knowlesi are the long-tailed macaque (Macaca fasicicularis), pig-tailed macaque (Macaca nemestrina), banded-leaf monkey (Presbytis melolophus)2 and stumped-tailed macaque (Macaca arctoides)3. Thus, the transmission of P. knowlesi is a complex interaction between the macaque hosts, the Anopheline vectors and humans. It has been hypothesised that urbanisation and deforestation has resulted in increased interactions between humans, macaques and the Anopheline vectors, accounting for the rise of P. knowlesi cases over the recent years4. Thus, it is important to be able to identify the prevalence of P. knowlesi in the macaque hosts as these are the major reservoirs of the parasite.
Apart from P. knowlesi, there are other Plasmodium species that are able to cause zoonotic infections in humans, four of which are found in wild macaques in Malaysia. The macaques, M. fascicularis and M. nemestrina, are known to harbour 5 Plasmodium species, which are P. knowlesi, Plasmodium cynomolgi, Plasmodium inui, Plasmodium coatneyi and Plasmodium fieldi. Since the first reported natural human infection of P. cynomolgi in 20145, there has been increasing reports of human P. cynomolgi infections throughout South East Asia, mostly as asymptomatic cases6,7,8,9,10,11. This suggests that P. cynomolgi could also be an emerging zoonotic threat, which needs to be further investigated. Recent reports have implied that P. inui too is capable of infecting humans under natural conditions12,13,14. Furthermore, surveillance studies using PCR-based methods in Thailand and indigenous communities in Malaysia, show that potentially P. coatneyi and P. fieldi could also be infecting humans12,14. Overall, the evidence suggests that all 5 of the common simian malaria parasites found in wild macaques in Malaysia should be considered zoonotic infectious agents in humans. Thus, it would be valuable to understand the prevalence of these species in the wild macaque population to be able to identify the zoonotic potential of these parasites.
The long-tailed macaque (Macaca fascicularis) is the most common macaque found in Peninsular Malaysia with an estimated population size of 133,403 in the year 201115. This species is also highly anthropophilic, resulting in increased interactions between M. fascicularis and humans. Microsatellite analysis in Borneo Malaysia have suggested that the majority of P. knowlesi human cases are caused by spill-over infections from the M. fascicularis population rather than the M. nemestrina population16. Thus, highlighting the importance of studying Plasmodium species in wild M. fascicularis populations.
Malaysia is divided into two regions, Peninsular Malaysia and Malaysia Borneo. A recent study has reported the prevalence of the 5 simian malaria parasites throughout Sarawak, Malaysia Borneo17. Although similar simian malaria surveillance studies have been conducted in Peninsular Malaysia, most have only focused on a small number of locations throughout Peninsular Malaysia18,19,20,21. Here, we determined the prevalence and distribution of P. knowlesi and the other simian malaria parasites in the M. fascicularis population throughout Peninsular Malaysia, by collecting M. fascicularis blood samples from 12 out of the 13 states in Peninsular Malaysia, which were then screened using a nested PCR approach.
Methods
Ethics and sample collection
Macaque blood samples were collected as part of a Wildlife Disease Surveillance Programme (WSDP), by the Department of Wildlife and National Parks (DWNP). The permission to conduct this research was obtained from Department of Wildlife and National Parks with the reference no. W-00256-15-19. The protocol for macaque sampling was approved by the University of California, Davis IACUC (Protocol number: 16048) and ethical approval for the use of macaque blood for malaria screening was approved by the Institutional Animal Care and Use Committee Universiti Malaya (UM IACUC) (Reference number: M/06122019/25022019-01/R). All methods were performed in accordance with relevant guidelines and regulations.
Long-tailed macaques were caught using baited traps and anesthetised using a ketamine/xylazine mixture (5 mg/kg of 100 mg/mL ketamine and 100 mg/mL xylazine) via the intramuscular route using a 21G needle by an attending veterinarian from the DWNP. The macaques were sexed and aged by the attending veterinarian, where the age of the macaques was determined by canine and nipple development in males and females respectively. Blood was withdrawn from the macaques with volumes ranging from 2 to 10 mL. This was based on the body weight of the macaques and blood was collected into lithium-heparin blood tubes using a 21G needle attached to a 5 mL syringe (Terumo, Philippines). Macaque blood samples were withdrawn at the trap sites. Whole blood samples were then frozen at − 20 °C until further use. This study was performed in concordance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
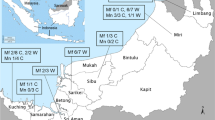
A minimum sample size of 384 macaques was estimated using the equation from Daniel 199922. In total, 410 M. fascicularis were caught in 12 states throughout Peninsular Malaysia (Fig. 1), with 62.1% (255/410) of the macaques being male and the majority of the macaques, 55.8% (229/410), being adults (Supplementary Table 1). All M. fascicularis sampled were wild macaques. Dates and respective sample sizes for each state are as follows:
Map of sampling locations (Left) with respective sample sizes (Right). Red dots show districts in which M. fascicularis were caught. The map of Peninsular Malaysia was created by the author using QGIS software version 3.6.3 with basemap shapefile modified from the original source on which the data had been plotted (https://data.humdata.org/dataset/cod-ab-mys).
Johor (n = 79, June 2019 to September 2020), Selangor (n = 63, October 2019 to June 2022), Pahang (n = 73, October 2019 to September 2022), Melaka (n = 5, November 2019), Kelantan (n = 31, February 2020), Kedah (n = 18, March 2020), Perak (n = 35, October 2020 to June 2022), Perlis (n = 6, February 2021), Negeri Sembilan (n = 19, August 2021 to September 2021), Terengganu (n = 59, June 2022 to September 2022), Kuala Lumpur (n = 19, June 2022 to September 2022), Putrajaya (n = 3, June 2022). For exact coordinates and sample sizes see Supplementary Data (ESRI: 102062 Kertau RSO Malaya Meters format).
DNA extraction
DNA was extracted from 100 μL of whole blood using the DNeasy® Blood and Tissue kit (QIAGEN) according to the manufacturer’s protocol. A total of 100 μL was eluted such that 1 μL of eluted DNA represented 1 μL of whole blood. DNA was stored at − 20 °C until further use. With each extraction batch, a DNA extraction negative control was added where 100 μL of phosphate buffered saline (PBS) was used as a sample.
Nested PCR assay
Four microliter of eluted DNA sample was used for screening. Samples were initially screened using a Plasmodium-specific nested PCR targeting the 18s rRNA gene as described previously23. Samples that were positive for Plasmodium parasites were then screened using a species-specific nested PCR targeting the 18s rRNA gene of P. knowlesi, P. cynomolgi, P. inui, P. coatneyi and P. fieldi as described previously24. Each batch of PCR included a DNA extraction negative control, a no template control and positive controls for each species. Positive controls came in the form of samples previously confirmed for each species with nested PCR20. For quality assurance, a set of random samples were taken and PCR repeated to confirm results. For representative gel images see Supplementary Figs. 1–3.
Results
Of the 410 M. fascicularis that were caught, 49.8% (204/410) were positive for malaria parasites. Pahang had the highest prevalence of malaria positive macaques, where 100% (73/73) were positive. This was followed by Johor (54.4%, 43/79), Terengganu (52.5%, 31/59), Selangor (49.2%, 31/63), Kelantan (48.4%, 15/31), Perak (25.7%, 9/35), Perlis (16.7%, 1/6) and Negeri Sembilan (5.3%, 1/19). All macaques caught from Melaka, Kedah, Kuala Lumpur and Putrajaya were negative (Table 1).
Throughout Peninsular Malaysia, P. inui was the most prevalent at 37.1%, followed by P. fieldi (32.9%), P. cynomolgi (31.2%), P. coatneyi (31.0%) and P. knowlesi (29.0%) (Table 2). It was noted that the predominant species varied between states where P. inui was the most prevalent for Johor, Pahang, Perak and Perlis, whilst P. cynomolgi was the most prevalent for Selangor, Kelantan and Negeri Sembilan (Table 2). The majority of infected macaques were infected by 2 or more Plasmodium species, where the most common infection type was an infection with all 5 simian Plasmodium species (Table 3). However, dual infections were the most common in Selangor, Kelantan, Perlis and Negeri Sembilan, whilst quadruple infections were the most common in Johor (Supplementary Table 2). This indicates that the species distribution and infection type of simian malaria parasites in the wild M. fascicularis population in Peninsular Malaysia is highly location dependant.
Discussion
Overall, this study determined the prevalence of the five simian malaria parasites in the wild M. fascicularis population of Peninsular Malaysia to be 49.8%. This is comparable to a previous study, with an earlier sampling period of 2016 to 2019, that looked at only seven states in Peninsular Malaysia and found a prevalence of 48.2%21. However, one limitation in comparing between these studies is that the methodology between the studies were not identical. The study conducted by Yusuf et al. performed PCR only on samples that were first confirmed positive via microscopy. Considering that microscopy is less sensitive than PCR, the prevalence of malaria in the M. fascicularis population could have been underestimated21. Notably, Pahang reported a prevalence of 100% with most of the macaques being infected with all 5 simian Plasmodium species. This indicates that Pahang is a potential hotspot for zoonotic malaria transmission and indeed, many of the P. knowlesi human cases in Peninsular Malaysia have been reported from Pahang25. Previous studies across a sampling period of 201620 and 2016 to 201921 reported a prevalence of 88.2% (30/34) and 93.6% (176/288) in Pahang, respectively. This could suggest that there could be an increasing trend of simian malaria prevalence in Pahang itself. Thus, although the overall trend of simian malaria prevalence throughout Peninsular Malaysia has not changed much over the years, there may be variations within specific states. This is evident in the differences found in species prevalence and infection types seen between the different states (Table 2, Supplementary Table 2). However, this study and previous studies investigating the prevalence of malaria in the macaque population have primarily been cross-sectional studies. Thus, it is difficult to infer this trend, which identifies the need for longitudinal-based studies.
Plasmodium knowlesi was the least prevalent (29.0%) of the 5 simian malaria species in the macaque population, but is the species responsible for the majority of human cases in Malaysia1. This highlights the complexity of zoonotic transmission to humans as different species may vary in transmissibility, exposure and susceptibility to humans. Spatial analysis of P. knowlesi human cases in Peninsular Malaysia from 2011 to 2018 revealed that P. knowlesi cases tended to cluster around the Kelantan-Pahang border, particularly in the Gua Musang (Kelantan) and Kuala Lipis (Pahang) districts25. Here we report a P. knowlesi macaque prevalence of 19.4% and 61.6% for Kelantan and Pahang, respectively. Notably, Johor (31.6%), Selangor (28.6%) and Terengganu (35.6%) reported a higher P. knowlesi prevalence than Kelantan. Thus, although prevalence of the parasite in the macaque host is an important risk factor26, this cannot be taken in isolation. Males with outdoor-based occupations such as plantation workers seem to be most at risk to P. knowlesi infection26. Furthermore, the tendency of P. knowlesi vectors to bite outdoors and have peak biting times earlier in the evening27 make interventions such as insecticide treated bed nets and indoor residual spraying ineffective to reduce P. knowlesi cases28. Thus, it would be important to combine data from the macaque population, mosquito behavioural studies and human cases in order to design a one health approach for interventions against P. knowlesi.
The prevalence of P. cynomolgi was high (> 30%) in Selangor, Pahang, Kelantan and in particular Terengganu, which is where the first reported P. cynomolgi human case was discovered in 20145. Since then, surveillance studies in Cambodia, Thailand and Malaysia have detected numerous accounts of P. cynomolgi cases in humans, most of which were asymptomatic8,11,12,14. Furthermore, case studies have reported P. cynomolgi infections in travellers returning from South East Asia9. This implies that further research into the epidemiology of P. cynomolgi is needed to ensure that P. cynomolgi does not become a major public health threat in the future. However, the identification of P. cynomolgi in patients can be difficult. This is due to the similarities between P. cynomolgi and P. vivax resulting in misidentification microscopically2 and in some cases even via molecular diagnosis5.
Plasmodium inui was the most prevalent species found in the wild M. fascicularis population (37.1%), with Pahang reporting a prevalence of 89.0%. This suggests that the risk of exposure of P. inui to humans could be high as P. inui was also the most prevalent species found in Anopheles mosquitoes sampled throughout Peninsular Malaysia29. Experimental infections with P. inui into humans have been shown to result in symptomatic febrile infections30 and a total of 24 human cases have been reported from Malaysia and Thailand12,13,14. The 5 human cases reported from Pahang and Melaka in Peninsular Malaysia and Sarawak in Malaysia Borneo were from surveillance studies rather than patients suggesting that these cases were all asymptomatic individuals12,13. Conversely, the 19 P. inui human cases reported from Thailand14, were all symptomatic patients reporting into a malaria clinic. However, 18/19 patients had coinfections with P. vivax and/or P. falciparum. Thus, it is difficult to identify whether P. inui contributed to the presentation of the symptoms. One patient was mono-infected with P. inui suggesting that P. inui infections in humans can result in symptomatic presentation, though rarely. Thus, it is possible that P. inui human infections may be currently underestimated due to the high likelihood of asymptomatic presentation. This highlights the need for large scale surveillance studies to identify the burden of P. inui infections in humans, especially in areas where P. inui is prevalent in the wild macaque hosts.
Evidence to suggest that P. coatneyi and P. fieldi infecting humans is still very preliminary. Although P. coatneyi and P. fieldi was found to be widely distributed across Peninsular Malaysia, previous experimental infections of P. coatneyi and P. fieldi to humans were unsuccessful2,31. However, two surveillance studies, have found P. coatneyi- and P. fieldi-positive patients via molecular techniques in Malaysia12 and Thailand14, respectively. For P. coatneyi, 3 patients from an indigenous community in Perak, Malaysia, were found to be positive. However, screening of the same samples in two different laboratories found inconsistent findings12. For, P. fieldi, 3 positive patients were found in Yala province in Southern Thailand, which borders Malaysia. Although these samples were microscopically positive, all three patients were coinfected with P. inui and P. vivax14. Overall, these studies provide preliminary evidence to suggest that P. coatneyi and P. fieldi can cause zoonotic infections in humans. Furthermore, these infections may be present at submicroscopic levels, suggesting screening using larger blood volumes, as has been found with both P. cynomolgi8 and P. inui13. Considering that the prevalence of P. coatneyi and P. fieldi in this study was 31.0% and 32.9% respectively, the zoonotic threat of these species in Peninsular Malaysia has yet to be determined.
One of the major limitations of this study is that samples were collected primarily through convenience sampling. Monkey baited traps were set up by the DWNP based on conflict case reports. This resulted in variable sampling periods across the states. Thus, one possibility is that the differences in prevalence between states could be due to temporal variations in transmission (e.g. during the monsoon vs dry seasons). This study has identified states of high prevalence such as Pahang (100%), Johor (54.4%), Terengganu (52.5%), Selangor (49.2%) and Kelantan (48.4%). Thus, to overcome the limitations due to temporal variation, these states could be used as sites for future longitudinal-based studies. Another limitation is that the reliance on convenience sampling has resulted in small sample sizes for Melaka (n = 5), Perlis (n = 6) and Putrajaya (n = 3). Thus, for these three states, it is difficult to conclude the prevalence of malaria in the M. fascicularis population. Thus, it would be beneficial for future studies to include these states with a higher sample size.
In conclusion, this study showed that the P. knowlesi, P. cynomolgi, P. inui, P. coatneyi and P. fieldi are widely distributed across Peninsular Malaysia in the M. fascicularis population. Although P. knowlesi was the least prevalent among the 5 species, it is the species that is responsible for the majority of symptomatic cases in humans25. In contrast, P. cynomolgi and P. inui although more prevalent in the macaque population, have comparatively fewer human cases and mostly result in asymptomatic infections8,11,12,13,14. This highlights that P. knowlesi is better adapted to grow in humans in comparison to the other simian malaria species2. Thus, an assessment of zoonotic risk should include both the prevalence in the macaque host as well as the adaptability of the parasite species to humans, among other factors. From a medical standpoint, all 5 simian malaria species can currently be treated with standard antimalarial drugs with no signs of drug resistance5,14. This is likely due to the absence of drug pressure on these populations in the main reservoir hosts, the macaques. As P. knowlesi is the main cause of malaria in Malaysia, residents and travellers to the different states should be made aware of the risk of potentially acquiring an infection in these states. Furthermore, as more human cases of P. cynomolgi, P. inui and potentially P. coatneyi and P. fieldi emerge, it would be beneficial for health care personnel to be aware of regions where these parasites are prevalent in the macaque population. This could help elucidate the zoonotic potential of these simian malaria species as zoonotic malaria may pose a threat to the goal of global malaria elimination.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
World Health Organization. WHO World Malaria Report 2023. Malaria Report 2023 (2023).
Coatney, G. R., Collins, W. E., Warren, M. & Contacos, P. G. The Primate Malarias (Department of Health Education and Welfare, 1971).
Fungfuang, W., Udom, C., Tongthainan, D., Kadir, K. A. & Singh, B. Malaria parasites in macaques in Thailand: Stump-tailed macaques (Macaca arctoides) are new natural hosts for Plasmodium knowlesi, Plasmodium inui, Plasmodium coatneyi and Plasmodium fieldi. Malar. J. 19, 1–7 (2020).
Fornace, K. M. et al. Association between landscape factors and spatial patterns of Plasmodium knowlesi infections in Sabah, Malaysia. Emerg. Infect. Dis. 22, 201 (2016).
Ta, T. H. et al. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar. J. 13, 1–7 (2014).
Putaporntip, C. et al. Plasmodium cynomolgi co-infections among symptomatic malaria patients, Thailand. Emerg. Infect. Dis. 27, 590 (2021).
Singh, B. et al. Naturally acquired human infections with the simian malaria parasite, Plasmodium cynomolgi, in Sarawak, Malaysian Borneo. Int. J. Infect. Dis. 73, 68 (2018).
Imwong, M. et al. Asymptomatic natural human infections with the simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J. Infect. Dis. 219, 695–702 (2019).
Hartmeyer, G. N. et al. Plasmodium cynomolgi as cause of malaria in tourist to Southeast Asia, 2018. Emerg. Infect. Dis. 25, 1936 (2019).
Grignard, L. et al. Natural human infections with Plasmodium cynomolgi and other malaria species in an elimination setting in Sabah, Malaysia. J. Infect. Dis. 220, 1946–1949 (2019).
Raja, T. N. et al. Naturally acquired human Plasmodium cynomolgi and P. knowlesi infections, Malaysian Borneo. Emerg. Infect. Dis. 26, 1801 (2020).
Yap, N. J. et al. Natural human infections with Plasmodium cynomolgi, P. inui, and 4 other simian malaria parasites, Malaysia. Emerg. Infect. Dis. 27, 2187 (2021).
Liew, J. W. et al. Natural Plasmodium inui infections in humans and Anopheles cracens mosquito, Malaysia. Emerg. Infect. Dis. 27, 2700 (2021).
Putaporntip, C. et al. Cryptic Plasmodium inui and Plasmodium fieldi infections among symptomatic malaria patients in Thailand. Clin. Infect. Dis. 75, 805–812 (2022).
Karuppannan, K. et al. Population status of long-tailed macaque (Macaca fascicularis) in Peninsular Malaysia. J. Primatol. 3, 34–37 (2014).
Divis, P. C. et al. Admixture in humans of two divergent Plasmodium knowlesi populations associated with different macaque host species. PLoS Pathog. 11, e1004888 (2015).
Nada-Raja, T. et al. Macaca fascicularis and Macaca nemestrina infected with zoonotic malaria parasites are widely distributed in Sarawak, Malaysian Borneo. Sci. Rep. 12, 10476 (2022).
Akter, R. et al. Simian malaria in wild macaques: First report from Hulu Selangor district, Selangor, Malaysia. Malar. J. 14, 1–9 (2015).
Ho, G. C. Prevalance of Plasmodium in the long-tailored macaque (MACACA FASICULARIS) from Selangor, Malaysia. In Oral Presentation at the 13th Association of Institutions for Tropical Veterinary Medicine (AITVM) Conference, Bangkok, Thailand, Vol. 13, 49 (2010).
Amir, A. et al. Natural Plasmodium infection in wild macaques of three states in Peninsular Malaysia. Acta Trop. 211, 105596 (2020).
Yusuf, N. M. et al. Plasmodium spp. in macaques, Macaca fascicularis, in Malaysia, and their potential role in zoonotic malaria transmission. Parasite 29, 1 (2022).
Naing, L., Winn, T. B. & Rusli, B. N. Practical issues in calculating the sample size for prevalence studies. Arch. Orofac. Sci. 1, 9–14 (2006).
Snounou, G. & Singh, B. Nested PCR analysis of Plasmodium parasites. Malaria Methods Protoc. 72, 189–203 (2002).
Lee, K. S. et al. Plasmodium knowlesi: Reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 7, e1002015 (2011).
Phang, W. K. et al. Spatial and temporal analysis of Plasmodium knowlesi infection in Peninsular Malaysia, 2011 to 2018. Int. J. Environ. Res. Public Health 17, 9271 (2020).
Grigg, M. J. et al. Individual-level factors associated with the risk of acquiring human Plasmodium knowlesi malaria in Malaysia: A case–control study. Lancet Planet Health 1, e97–e104 (2017).
Vythilingam, I., Wong, M. L. & Wan-Yussof, W. S. Current status of Plasmodium knowlesi vectors: A public health concern? Parasitology 145, 32–40 (2018).
William, T. et al. Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax malaria in Sabah, Malaysia. PLoS Negl. Trop. Dis. 7, e2026 (2013).
Jeyaprakasam, N. K. et al. High transmission efficiency of the simian malaria vectors and population expansion of their parasites Plasmodium cynomolgi and Plasmodium inui. PLoS Negl. Trop. Dis. 17, e0011438 (2023).
Coatney, G. R., Chin, W., Contacos, P. G. & King, H. K. Plasmodium inui, a quartan-type malaria parasite of Old World monkeys transmissible to man. J. Parasitol. 52, 660–663 (1966).
Contacos, P. G. & Coatney, G. R. Experimental adaptation of simian malarias to abnormal hosts. J. Parasitol. 49, 912–918 (1963).
Acknowledgements
This study was funded by the Ministry of Higher Education Malaysia, Long Term Research Grant Scheme (LRGS/1/2018/UM/01/1/2). The permission to conduct this research was obtained from Department of Wildlife and National Parks with the reference no. W-00256-15-19.
Author information
Authors and Affiliations
Contributions
A.A., F.M.Y., W.M.M.A. and L.Y.L. conceived the project. S.S. and A.A. conceptualised and planned experiments. M.L.A., A.A.I.R. and N.A. was involved in macaque sample collection. S.S. prepared, managed and performed experiments. S.S. and A.A. managed ethical clearance. S.S., M.L.A. and A.A. compiled, analysed and interpreted the data. S.S. prepared the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shahari, S., Bin Abdullah, M.L., Binti Isman Rohimly, A.A. et al. The prevalence of simian malaria in wild long-tailed macaques throughout Peninsular Malaysia. Sci Rep 14, 6023 (2024). https://doi.org/10.1038/s41598-024-54981-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-54981-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.