Abstract
Although chorioamnionitis (CAM) has been demonstrated to be associated with numerous short- and long-term morbidities, the precise mechanisms remain unclear. One of the reasons for this is the lack of appropriate models for analyzing the relationship between the fetal environment and chorioamnionitis and fetal programming in humans. In this study, we aimed to clarify the fetal programming caused by CAM using the gene expression profiles of UCMSCs. From nine preterm neonates with CAM (n = 4) or without CAM (n = 5), we established UCMSCs. The gene expression profiles obtained by RNA-seq analysis revealed distinctive changes in the CAM group USMSCs. The UCMSCs in the CAM group had a myofibroblast-like phenotype with significantly increased expression levels of myofibroblast-related genes, including α-smooth muscle actin (p < 0.05). In the pathway analysis, the genes involved in DNA replication and G1 to S cell cycle control were remarkably decreased, suggesting that cellular proliferation was impaired, as confirmed by the cellular proliferation assay (p < 0.01–0.05). Pathway analysis revealed that genes related to white fat cell differentiation were significantly increased. Our results could explain the long-term outcomes of patients who were exposed to CAM and revealed that UCMSCs could be an in vitro model of fetal programming affected by CAM.
Similar content being viewed by others
Introduction
Fetal programming occurs during embryonic and fetal development, and may cause persistent effects on the fetus and infant long after birth. The concept of fetal programming is derived from the fetal origins hypothesis, that is, Barker’s hypothesis. The concept of fetal programming has established a new approach to identifying the causes of disease, shifting to the in utero environment and its critical role in lifelong health. Fetal programming has been presumed to affect individual gene expression by epigenetic modifications, in which genes are expressed differently without any change to the DNA sequence itself. Further, other biological processes have been supposed to be involved, such as changes in molecular biological functions to permanent hormonal changes, alterations in metabolism, or responses to physiological stressors1.
Fetal undernutrition is the most important factor affecting fetal programming, and it has been intensively investigated to date. The fetus adapts to the intrauterine nutritional environment through changes in the regulation of metabolism and redistribution of blood flow, controlling fetal growth2. Indeed, the birth weight of babies born after oocyte donation is strongly associated with the weight of the recipient mother rather than the weight of the donor1. Fetal nutritional adaptation is explained by the thrifty phenotype hypothesis, and fetal growth restriction has been shown to increase the risk for lifestyle diseases in adults, such as cardiovascular disease, obesity, and diabetes.
Chorioamnionitis is defined as the presence of active infection in the amniotic sac that causes inflammatory changes in the mother. Chorioamnionitis is associated with numerous short-term and long-term morbidities, such as intraventricular hemorrhage, retinopathy of prematurity, chronic lung disease, and cerebral palsy3. To date, several studies using animal models have suggested that chorioamnionitis affects fetal programming4; however, precise mechanisms are mostly not clarified, especially chorioamnionitis in humans. This is due to the lack of appropriate models for analyzing the relationship between fetal inflammatory environments and fetal programming.
Mesenchymal stem cells (MSCs) have the capacity for proliferation, multilineage differentiation, and immunomodulatory properties, and the properties of cultured MSCs in vitro have been suggested to be applicable for broad medical applications such as regenerative medicine and immunomodulatory medicine against graft versus host disease (GVHD)5 and SLE6. As a source of MSCs, umbilical cord tissue has been considered a promising option for several reasons. First, the umbilical cord is traditionally regarded as a waste tissue, so isolating MSCs from umbilical cord tissue is not ethically controversial compared with obtaining MSCs from other tissues, such as bone marrow. Second, UCMSCs are mainly located in the subcortical endothelium of the umbilical cord, perivascular region, and Wharton’s jelly (WJ), and can be isolated from umbilical cords by explant monolayer culture, which does not require complicated procedures. Third, in UCMSCs, the expression of human leukocyte antigen (HLA) -ABC and HLA-DR is low, resulting in lower alloreactivity.
In addition to the utilities in regenerative medicine, recent reports suggest that the UCMSCs may be programmed in a manner similar to other MSCs in the fetus, and would be an in vitro model for fetal growth restriction7. Analyses of UCMSCs from the neonates born small for gestational age (SGA) were useful in identifying pathways specific to fetal growth restriction7. In UCMSCs from neonate with fetal growth restriction, early growth response 1 (EGR1) and cyclooxygenase 2 (Cox2) in the UCMSCs are affected, leading to lower insulin sensitivity and increased adipogenicity7.
In the present study, to understand fetal programming caused by CAM, we analyzed UCMSCs obtained from preterm neonates with a history of CAM. Our analysis of USMSCs revealed that UCMCSs could be an in vitro model of fetal programming affected by CAM, providing a tool for clarifying the molecular mechanisms of postnatal complications in neonates who experienced CAM during the fetal period.
Materials and methods
Subjects and samples
The present study was approved by the ethical board of Tokyo Medical and Dental University Graduate School of Medicine (M2017-28) and conducted in accordance with the approved guidelines. Written informed consent was obtained from the parents of each neonate. Human umbilical cords were collected from nine very low birth weight infants delivered by cesarean sections at 25–30 weeks of gestation. Clinical data were prospectively collected from the medical records of the neonates and their mothers.
We divided the nine neonates into CAM (n = 4) and non-CAM groups (n = 5). The neonates who exhibited “Triple I,” namely intrauterine inflammation, or infection, or both, were categorized into the CAM group. More specifically, besides pathological findings, maternal fever (> 38.0C), leukocytosis (> 15,000), fetal tachycardia (> 160/min), and definite purulent fluid from the cervical os, were considered as symptoms of CAM8. Further, we confirmed CAM by histological analysis of the placenta. On the other hand, with or without identifying histological CAM in placentas, asymptomatic cases were categorized into non- CAM group.
The criterion for chronic lung disease was the requirement of oxygen support at 36 weeks’ corrected postnatal gestational age9. Neuromotor development was evaluated according to the Kyoto Scale of Psychological Development 2001, a developmental test that has been widely used by Japanese clinicians working with infants, toddlers, and children. We classified the subjects into three groups based on the TDQ score (normal: > 85, border: 70 ~ 85 and retardation: < 70) (Shinpan K Shiki Hattatsu Kensahou 2001 Nenban)10.
Preparation of UCMSCs
Umbilical cord-derived mesenchymal stem cells (UCMSCs) were established according to an improved explant method previously reported11. Briefly, a small fragment of the umbilical cord was cultured at 37 °C (5% CO2 and 95% air) in MEM-α (Thermo Fisher Scientific, Waltham, MA, USA) with 10% FBS and 2% penicillin–streptomycin (Thermo Fisher Scientific). The outgrowth monolayer cells (Passage1: P1) were collected by disassociating with TrypLE™ Express enzyme (Thermo Fisher Scientific). The collected cells were seeded into the new dishes and frozen stock was collected after reaching confluence (P2). In the present study, we used the cells from the freeze stock (P3).
RNA extraction
Total RNA from UCMSCs was extracted and purified using the RNeasy Micro Kit (#74106, Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA concentration was measured using a Nanodrop ND-8000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
Cell surface marker analysis
UCMSCs were dissociated with TrypLE™ Express enzyme (Thermo Fisher Scientific), washed with PBS and suspended. The cells were incubated with phycoeryhrin- (PE-) or Fluoresceinisothiocyanate isomer-I (FITC-) conjugated mouse primary antibodies against CD14, CD19, CD34, CD45, CD73, CD90, CD105, or HLA-DR (BD Bioscience, Franklin Lakes, NJ) for 10 min at room temperature and washed with PBS. Flow cytometry and analysis was performed using BD LSRFortessa™, FACSDiva software and FlowJo™ software (BD Bioscience).
RNA seq
Library preparation and sequencing
The sequencing libraries from total RNA of USMSC were constructed using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (#E7760, New England Biolabs, MA, USA) with NEBNext Poly(A) mRNA Magnetic Isolation Module according to the manufacturer’s protocols. The quality of the libraries was assessed using an Agilent 2200 TapeStation High Sensitivity D1000 (Agilent Technologies, Inc., Santa Clara, CA, USA). The pooled libraries of the samples were sequenced using the Illumina NextSeq 500 (Illumina, Inc., San Diego, CA, USA) in 76-base-pair (bp) single-end reads.
Alignment to the whole transcriptome
Sequencing adaptors, low quality reads, and bases were trimmed using the Trimmomatic-0.38 tool12. The sequence reads were aligned to the human reference genome (hg19) using STAR 2.713. For the whole transcriptome alignment with the STAR, files of the gene model annotations and known transcripts were downloaded from the Illumina’s iGenomes website (http://support.illumina.com/sequencing/sequencing_software/igenome.html).
Quantifying the gene expression levels and detection of differentially expressed genes
The aligned reads were subjected to downstream analyses using StrandNGS 3.2 software (Agilent Technologies). The read counts allocated for each gene and transcript RefSeq Genes (2015.10.05) were quantified using the trimmed mean of M-value (TMM) method14. To investigate gene expression differences, we selected genes through moderated t-test (Benjamini–Hochberg multiple test correction FDR-q-value < 0.05) and up- or downregulated them by setting a threshold of twofold. To summarize the biological aspects of the selected genes, we employed a volcano plot, Gene Ontology (GO) terms, and pathway analysis.
For the volcano plot, the genes of each category were selected using the following procedures. The representative genes involved in the contractile apparatus and extracellular matrix were selected based on previous reports15,16,17,18. We selected cell cycle genes that annotated GO:0045787 positive regulation of cell cycle, fold change < 0.5, and p < 0.05, or that annotated GO:0045786 negative regulation of cell cycle, fold change > 2, and p < 0.05. We used R software version 4.1.1 (R-Tools Technology Inc., ON, Canada) for statistical analysis.
Pathway statistical analysis was performed on a pathway collection of the WikiPathways19 database using PathVisio tool20 to determine pathways containing the most changed expression, taking into consideration the number of genes in the pathway that were measured in the experiment and the number of genes that were differentially expressed.
Quantitative real-time PCR
cDNA was synthesized from 800 ng of total RNA from UCMSCs by using a High Capacity cDNA Reverse Transcription Kit (#4368814, Thermo). Real-time PCR analysis was performed with a Roche Lightcycler 480II real-time PCR system (Roche Diagnostics, Mannheim, Germany) using FastStart Universal SYBR Green master mix (#4913914001, Roche) with 0.5 μM sense and antisense primers and cDNA (corresponding to 25 ng total RNA) according to the manufacturer’s instructions. The relative expression of each transcript was calculated based on the calibration curve method using GAPDH as an endogenous reference for normalization. The primer sets are listed in Table S2. Biologically independent (n = 4 or 5) experiments were performed, and all sample measurements were repeated at least three times.
Cell proliferation assay
UCMSCs were seeded at the density of 1 × 106 cells per 10 cm cell culture dish. The cells were passaged three times every 2 days and the number of the cells at each passage was counted. Biologically independent (n = 4 or 5) experiments were performed, and all sample measurements were repeated at least twice.
MTS assay
At the density of 1 × 104 cells per 96 well plate, we seeded UCMCSs, and the cells were incubated at 37 °C (5% CO2 and 95% air) for 24 h. Cell proliferation was measured by the CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (#G3582, Promega, Madison, WI, USA) according to the manufacturer’s instruction. Briefly, 20 μl of MTS reagent (a tetrazolium compound and an electron coupling reagent) was added into each well and incubated at 37 °C (5% CO2 and 95% air) for 4 h. The absorbance at 450 nm was measured using an iMark™ Microplate Reader (#168-1130JA, BIO-RAD Laboratories, Inc, Hercules, CA). Biologically three independent experiments were performed, and all sample were measured with five replicates. We calculated the average and SE of each sample.
Cell cycle analysis
UCMSCs were seeded at 6 × 105 cells/per 10 cm dish and incubated at 37 °C (5% CO2 and 95% air) for 24 h. At 60–70% confluence, cells were performed cell cycle analysis. Briefly, cells were washed and fixed in 70% ethanol for 2 h at – 20 °C. Fixed cells were washed and incubated in 0.25 mg/ml RNase A (#12091039, Thermo Fisher Science) for 30 min at 37 °C. Subsequently, cells were stained with 50 μg/ml propidium iodide (PI) (#25535-16-4, BioVison, Inc, Milpitas, CA) for 30 min at 4 °C in the dark. Cell cycles were assessed by flow cytometry and analysis was performed using BD LSRFortessa™, FACSDiva software and FlowJo™ software (BD Bioscience), and counted the number of the cells at each cell cycle phase.
Statistical analysis
Real-time PCR was analyzed using the Mann–Whitney U test. Student’s t test was used for cell proliferation and cell cycle analyses. Clinical data and experimental data were compared using Fisher’s exact test, Mann–Whitney U test, or Student’s t test, as required. Cell cycle analysis.
For statistical analysis, we used JMP Pro version 15.1.0 (SAS Institute Inc, NC, USA). Statistical significance was set at p < 0.05. Significant differences were expressed as ‘*’ for P values, 0.05, ‘**’ for P values ,0.01 and ‘***’ for P-values ,0.001 respectively.
Results
The UCMSCs would be an in vitro model of fetal programming affected by CAM
We obtained the cells from umbilical cords of neonates with CAM (n = 4) and normal healthy controls (n = 5) by explant monolayer culture. There were no significant differences in the clinical backgrounds between the normal healthy group and the CAM group, except for the pathological grade of chorioamnionitis and funisitis (Table 1). No significant differences in the frequency of CLD and neurodevelopment until 3 years old between the two groups (Supplementary Table 1) were observed. We selected three samples from each group and performed transcriptome analysis.
The collected cells from umbilical cords highly expressed CD73, CD90 and CD105, while the expression of CD34, CD45, CD14, CD19, and HLA-DR were low, indicating that the cells had the features of UCMSCs (Fig. 1A, Fig. S1). On the other hand, principal component analysis revealed that the gene expression profiles were distinctively changed in the CAM group (Fig. 1B), and a substantial number of genes were significantly upregulated or downregulated (Fig. 1C, Table 2).
(A): Surface marker analysis by flow cytometry revealed that the cells obtained from umbilical cords had UCMSC features. The cells expressed CD73, CD90, and CD105, while not CD34, CD45, CD14, CD19, and HLA-DR; (B): Principal component analysis revealed that the gene expression profiles of the CAM group (red) were distinctively different from the normal healthy control, non-CAM group (yellow). (C): Volcano plot displaying the results of RNA-seq results from the CAM group and normal healthy controls. The genes that passed the thresholds for p value (Log10 1/p value > 1.30) and log fold change (Log2 fold change > + 1 or < −1) were colored (red for upregulated and blue for downregulated).
The features of the gene expression profile of UCMSCs in the CAM group
Myofibroblast-like profiles
Gene ontology analyses revealed that the genes annotated to extracellular structure, collagen catabolic process and collagen metabolic process were remarkably activated (Table 3). Furthermore, pathway analysis showed that the genes associated with focal adhesion, striated muscle contraction, and human primary endometrial stromal cells were significantly upregulated (Table 4). This observation supported the data from the gene expression profile in which the myofibroblast-related genes were upregulated (Fig. 2A). Quantitative RT-PCR analysis confirmed that the expression level of the myofibroblast marker, α-smooth muscle actin (α-SMA) was significantly increased (Fig. 2B), suggesting that the phenotypes of UCMSCs with CAM would be shifted to the that of myofibroblasts.
(A): Genes related to the contractile apparatus (light blue) and extracellular matrix (deep blue), were plotted on the volcano plot. (B): Real time analysis of the myofibroblast marker genes, the CAM group: n = 4, and the non CAM group (CTRL): n = 5, *p < 0.05, Error bars indicate ± SE, Mann–Whitney analysis was used for statistical analysis.
Inhibition of the cell cycle
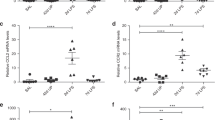
The cell cycle of UCMSCs from the CAM group was significantly suppressed (Table 3). The downregulation of the genes annotated to activate cell cycles, such as cell cycle, mitotic cell cycle, DNA replication, DNA-dependent DNA replication, etc., were statistically significant (Fig. 3A, Table 3). The cell cycle suppressor genes, including genes involved in cell cycle arrest, were activated (Fig. 3A, Table 3). Pathway analysis revealed that the genes involved in DNA replication, G1 to S cell cycle control, cell cycle, and ATM signaling pathway were remarkably decreased (Table 4). Consistently, the proliferation of UCMSCs in the CAM group was significantly impaired (Fig. 3B,C), and the population in the synthesis (S), and the second growth (G2) phases were reduced in the CAM group (Fig. 3D,E).
(A): Genes related positive regulation of cell cycle (red), and negative regulation of cell cycle (blue) were plotted on the volcano plot. (B): Cell proliferation assay. We passaged the cells three times every 2 days (days 2, 4, and 6) and counted the number of the cells at each passage. (control: n = 5, CAM: n = 4) *p < 0.05, **p < 0.01, Error bars indicate ± SE. Student’s t test was used for statistical analysis. (C): MTS assay of UCMSCs statistically revealed the significant difference between the control (n = 3) and the CAM group (n = 3). *p < 0.05, Error bars indicate ± SE. Student’s t test was used for statistical analysis. (D): Representative data of the cell cycle distribution of the UCMSCs from the control and the CAM group. Compared with the control group, the cells in G2/S phase were decreased in the CAM group. (E): A comparison of the UCMSCs population at each phase of the cell cycle revealed the significant difference between the control (n = 4) and the CAM group (n = 4), ***p < 0.001, Error bars indicate ± SE. Student’s t test was used for statistical analysis.
Activated adipogenesis related genes
In pathway analysis, the genes related to adipogenesis, including transcription factor regulation in adipogenesis, white fat cell differentiation, and leptin insulin overlap were significantly increased (Table 4). In contrast, brown fat cell differentiation related genes did not change significantly (data not shown). This observation suggests that CAM during the fetal period selectively activates the genes that contribute to the differentiation of white fat cells.
Discussion
Our analyses revealed that a fetal environment with CAM affected the characteristics of MSCs with dramatically altered gene expression profiles, promoting myofibroblastic and white adipocyte differentiation with reduced capability for cell proliferation. Our present study highlights three points: (a) UCMSCs would be an excellent model that reflect the fetal programming due to CAM, (b) the affected gene expression profiles of UCMSC by CAM could explain the postnatal complications of neonates who experienced CAM during the fetal period, and (c) for future utilization of autologous cell treatment, CAM would affect the outcomes of the therapy.
Our analysis revealed that the genetic expression profiles of UCMSC were profoundly deviated by exposure to CAM, suggesting that CAM would cause fetal programming to affect the long- term outcomes of offspring. In the context of fetal programming, extensive research has focused on maternal nutritional status and later metabolic disease in neonates, and CAM has been a sub-focus. Indeed, the major adverse outcomes due to CAM, such as sepsis and neurodevelopmental problems, are mainly caused by the direct effects of infection or inflammatory injury due to cytokines21,22. However, in the last decade, the placenta has become a new focus in fetal programming research23. If placental function, which underpins fetal development, is impaired, fetal development may be compromised. Historically, one of the most striking fetal programming findings come from examination of the subjects who were exposed to the 1918 (Spanish) influenza pandemic in utero24. Accordingly, it is not surprising that CAM, a condition of placental inflammation, would lead to compromised fetal programming. Additionally, we consider the possibility that infection or inflammatory injury could be another cause for fetal programming. For clarifying the possibility, further accumulation of data from the cases and disease models are required.
Recently, programmed differences in UCMSCs have been reported to reflect the effects of a maternal metabolic environment, and UCMSC is a model of fetal programming7,25. One of the peculiar characteristics of UCMCSs from CAM neonates was a myofibroblast-like phenotype. Myofibroblasts are contractile, α-smooth muscle actin-positive cells with multiple roles in pathophysiological processes including mediating wound contraction26. The persistent presence of myofibroblasts in tissues promotes tissue fibrosis. CLD, also known as bronchopulmonary dysplasia (BPD), is the most common chronic respiratory disorder in preterm infants and is characterized by an interruption in pulmonary vascular and alveolar development27. The etiology of CLD is multifactorial and involves antenatal and/or postnatal factors, such as intrauterine growth restriction, maternal smoking, mechanical ventilation, oxygen toxicity, and infection, which impair lung maturation. Although it is not conclusive, some studies have revealed an association between CAM and CLD8,28,29,30, and MSC differentiation into myofibroblasts is likely to be involved in the pathophysiology of CLD17. Our data, the myofibroblastic phenotypes of UCMSCs may explain the risk of CLD in neonates with CAM.
Our analysis found that s from CAM patients highly expressed white adipocyte related genes, suggesting that CAM alters the programmed gene expression profile in MSC metabolism. As previously observed in the influence of maternal metabolic environment on the fetal metabolome and genome in7,31, CAM also might cause programmed differences in stem cell metabolism, which could lead to differences in body composition in later life stages. Although the association between CAM and body composition or lipid metabolism in later life has not been clarified, prenatal inflammation of the placenta would cause metabolic disorders, as described in cases of exposure to the 1918 influenza pandemic during the fetal period32.
The proliferative ability of UCMSCs was profoundly affected by CAM. This suggests that the proliferative ability of USMSCs is vulnerable to the inflammatory environment during the fetal period. MSCs are thought to directly respond to inflammatory stimuli by cytokine priming, leading to acquiring their anti-inflammatory and immunomodulatory activities in situ33,34,35. The anti-inflammatory and immunomodulatory effects can occur through cellular contact and/or the secretion of diverse factors36, and the characteristics of MSCs enable immune modulation and anti-inflammatory applications that are broadly applicable in damaged tissue. Although, the significance of cellular growth for the therapeutic applications has not been clarified, our data suggest that inflammatory stimuli during the fetal period potentially affect the outcomes of therapeutic applications with UCMSCs. In addition to UCMSCs, the profiles of other neonate derived MSCs, such as cord-blood derived MSCs, could be affected by inflammatory stimuli during the fetal period. Based on our findings, further careful evaluation of the influence of inflammatory stimuli on neonate derived MSCs is required.
The present study has some limitations. Our findings are mainly based on the gene expression profiles of UCMSCs, and the biological and clinical relevance of these findings have not been clarified in detail. Despite this limitation, we presume that UCMSCs is a potential mode of the fetal programming caused by CAM. The characteristics of the gene expression profiles in UCMSCs from CAM neonates could explain the pathophysiology of the complications due to CAM, such as chronic lung disease (CLD). Long-term follow-up and trajectory analysis of CAM cases with a large cohort is essential to elucidate the biological and clinical relevance of UCMSC as a model of a fetal inflammatory environment.
In summary, our data revealed that UCMSC would be an excellent model that reflects fetal programming, and CAM is another factor that causes fetal programming, affecting the long-term outcomes of offspring. Our findings would provide valuable insights for understanding fetal programming caused by CAM and the development of optimal protocols for the applications of regenerative medicine using USMSCs.
References
Cunningham, S. & Cameron, I. T. Consequences of fetal growth restriction during childhood and adult life. Curr. Obstet. Gynaecol. 13, 212–217. https://doi.org/10.1016/s0957-5847(03)00039-8 (2003).
Fowden, A. L. Endocrine regulation of fetal growth. Reprod. Fertil. Dev. 7, 351–363. https://doi.org/10.1071/rd9950351 (1995).
Polam, S., Koons, A., Anwar, M., Shen-Schwarz, S. & Hegyi, T. Effect of chorioamnionitis on neurodevelopmental outcome in preterm infants. Arch. Pediatr. Adolesc. Med. 159, 1032–1035. https://doi.org/10.1001/archpedi.159.11.1032 (2005).
Chin, P. Y. et al. Toll-like receptor-4 antagonist (+)-naloxone confers sexually dimorphic protection from inflammation-induced fetal programming in mice. Endocrinology 160, 2646–2662. https://doi.org/10.1210/en.2019-00493 (2019).
Muller, I. et al. Application of multipotent mesenchymal stromal cells in pediatric patients following allogeneic stem cell transplantation. Blood Cells Mol. Dis. 40, 25–32. https://doi.org/10.1016/j.bcmd.2007.06.021 (2008).
Sun, L. et al. Umbilical cord mesenchymal stem cell transplantation in severe and refractory systemic lupus erythematosus. Arthritis Rheum 62, 2467–2475. https://doi.org/10.1002/art.27548 (2010).
Sukarieh, R. et al. Molecular pathways reflecting poor intrauterine growth are found in Wharton’s jelly-derived mesenchymal stem cells. Hum. Reprod. 29, 2287–2301. https://doi.org/10.1093/humrep/deu209 (2014).
Peng, C. C., Chang, J. H., Lin, H. Y., Cheng, P. J. & Su, B. H. Intrauterine inflammation, infection, or both (Triple I): A new concept for chorioamnionitis. Pediatr. Neonatol. 59, 231–237. https://doi.org/10.1016/j.pedneo.2017.09.001 (2018).
Shennan, A. T., Dunn, M. S., Ohlsson, A., Lennox, K. & Hoskins, E. M. Abnormal pulmonary outcomes in premature infants: Prediction from oxygen requirement in the neonatal period. Pediatrics 82, 527–532 (1988).
Society for the Kyoto Scale of Psychological Development Test. Shinpan K Shiki Hattatsu Kensahou 2001 Nenban [The Kyoto Scale of Psychological Development Test 2001]. (Nakanishiya Shuppan, Kyoto, 2008 (in Japanese)).
Mori, Y. et al. Improved explant method to isolate umbilical cord-derived mesenchymal stem cells and their immunosuppressive properties. Tissue Eng. Part C Methods 21, 367–372. https://doi.org/10.1089/ten.TEC.2014.0385 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25. https://doi.org/10.1186/gb-2010-11-3-r25 (2010).
Avery, D. et al. Extracellular matrix directs phenotypic heterogeneity of activated fibroblasts. Matrix Biol. 67, 90–106. https://doi.org/10.1016/j.matbio.2017.12.003 (2018).
Klingberg, F., Hinz, B. & White, E. S. The myofibroblast matrix: Implications for tissue repair and fibrosis. J. Pathol. 229, 298–309. https://doi.org/10.1002/path.4104 (2013).
Popova, A. P. et al. Autocrine production of TGF-beta1 promotes myofibroblastic differentiation of neonatal lung mesenchymal stem cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 298, L735-743. https://doi.org/10.1152/ajplung.00347.2009 (2010).
Huang, J. et al. Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J. Clin. Invest. 118, 515–525. https://doi.org/10.1172/jci33304 (2008).
Slenter, D. N. et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 46, D661-d667. https://doi.org/10.1093/nar/gkx1064 (2018).
Kutmon, M. et al. PathVisio 3: An extendable pathway analysis toolbox. PLoS Comput. Biol. 11, e1004085. https://doi.org/10.1371/journal.pcbi.1004085 (2015).
Jobe, A. H. Effects of chorioamnionitis on the fetal lung. Clin. Perinatol. 39, 441–457. https://doi.org/10.1016/j.clp.2012.06.010 (2012).
Cordeiro, C. N., Tsimis, M. & Burd, I. Infections and brain development. Obstet. Gynecol. Surv. 70, 644–655. https://doi.org/10.1097/OGX.0000000000000236 (2015).
Burton, G. J., Fowden, A. L. & Thornburg, K. L. Placental origins of chronic disease. Physiol. Rev. 96, 1509–1565. https://doi.org/10.1152/physrev.00029.2015 (2016).
Helgertz, J. & Bengtsson, T. The long-lasting influenza: The impact of fetal stress during the 1918 influenza pandemic on socioeconomic attainment and health in Sweden, 1968–2012. Demography 56, 1389–1425. https://doi.org/10.1007/s13524-019-00799-x (2019).
Baker, P. R. 2nd. et al. Altered gene expression and metabolism in fetal umbilical cord mesenchymal stem cells correspond with differences in 5-month-old infant adiposity gain. Sci. Rep. 7, 18095. https://doi.org/10.1038/s41598-017-17588-4 (2017).
Pakshir, P. et al. The myofibroblast at a glance. J. Cell. Sci. https://doi.org/10.1242/jcs.227900 (2020).
Hwang, J. S. & Rehan, V. K. Recent advances in bronchopulmonary dysplasia: Pathophysiology, prevention, and treatment. Lung 196, 129–138. https://doi.org/10.1007/s00408-018-0084-z (2018).
Villamor-Martinez, E. et al. Association of chorioamnionitis with bronchopulmonary dysplasia among preterm infants: A systematic review, meta-analysis, and metaregression. JAMA Netw. Open 2, e1914611. https://doi.org/10.1001/jamanetworkopen.2019.14611 (2019).
Tang, Q., Zhang, L., Li, H. & Shao, Y. The fetal inflammation response syndrome and adverse neonatal outcomes: A meta-analysis. J Matern Fetal Neonatal Med 34, 3902–3914. https://doi.org/10.1080/14767058.2019.1702942 (2021).
Mittendorf, R. et al. Special relationships between fetal inflammatory response syndrome and bronchopulmonary dysplasia in neonates. J. Perinat. Med. 33, 428–434. https://doi.org/10.1515/jpm.2005.076 (2005).
Joseph, R. et al. ACSL1 is associated with fetal programming of insulin sensitivity and cellular lipid content. Mol Endocrinol 29, 909–920. https://doi.org/10.1210/me.2015-1020 (2015).
Mazumder, B., Almond, D., Park, K., Crimmins, E. M. & Finch, C. E. Lingering prenatal effects of the 1918 influenza pandemic on cardiovascular disease. J Dev Orig Health Dis 1, 26–34. https://doi.org/10.1017/s2040174409990031 (2010).
Kim, S. H. et al. Transcriptome sequencing wide functional analysis of human mesenchymal stem cells in response to TLR4 ligand. Sci. Rep. 6, 30311. https://doi.org/10.1038/srep30311 (2016).
Ryan, J. M., Barry, F., Murphy, J. M. & Mahon, B. P. Interferon-gamma does not break, but promotes the immunosuppressive capacity of adult human mesenchymal stem cells. Clin. Exp. Immunol. 149, 353–363. https://doi.org/10.1111/j.1365−2249.2007.03422.x (2007).
Krampera, M. et al. Role for interferon-gamma in the immunomodulatory activity of human bone marrow mesenchymal stem cells. Stem Cells 24, 386–398. https://doi.org/10.1634/stemcells.2005-0008 (2006).
Aggarwal, S. & Pittenger, M. F. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood 105, 1815–1822. https://doi.org/10.1182/blood-2004-04-1559 (2005).
Acknowledgements
The authors thank Kazue Kinoshita from Molecular laboratory, Tokyo Metropolitan Children's Medical Center for supporting experiments and Xiaohui Song from Department of Pediatrics and Developmental Biology, Tokyo Medical and Dental University.
Funding
This Research was supported by Clinical Research Fund of Tokyo Metropolitan Government #R01050301 and Japan Society for the Promotion of Science (JSPS) Kakenhi (Grant Number: 20K22868, 15K09708).
Author information
Authors and Affiliations
Contributions
Y.N., A.T., K.K., and T.M. designed the research. Y.N. and A.T. performed experiments. Y.N, A.T., I.H., M.S., T.S., K.I, H.I, C.M. treated patients, collected samples, clinical data and performed analysis. Y.N. and K.K. wrote the manuscript. A.K., K.O., M.K., T.M. critically reviewed the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Noguchi, Y., Taki, A., Honda, I. et al. Transcriptome analysis of umbilical cord mesenchymal stem cells revealed fetal programming due to chorioamnionitis. Sci Rep 12, 6537 (2022). https://doi.org/10.1038/s41598-022-10258-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10258-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.