Abstract
Response to anti-TNF therapy is of pivotal importance in patients with Crohn’s disease (CD). Here we integrated our and previously reported PBMC derived transcriptomic and genomic data for identification of biomarkers for discrimination between responders and non-responders to anti-TNF therapy. CD patients, who were naïve with respect to the treatment with biologicals, were enrolled in the study. DNA and RNA were extracted from peripheral blood mononuclear cells. RNA-seq was performed using BGISEQ-500. Genotyping was performed using Infinium Global Screening Array. Association regressions were carried out with 12 week response to adalimumab as an outcome variable. RNA-seq analysis confirmed 7 out of 65 previously suggested genes involved in anti-TNF response. Subsequently, analysis of single nucleotide variants in regions of confirmed genes identified 5 variants near MMD and two in ELOVL7 intronic regions associated with treatment response to anti-TNF. Functional analysis has shown that rs1465352, rs4422035 and rs78620886 are listed at H3K9ac_Pro histone modification epigenetic mark. The present study confirmed MMD and ELOVL7 involvement in anti-TNF response and revealed that the regulation of MMD and ELOVL7 gene regions in ADA response may be a part of a complex interplay extending from genetic to epigenetic and to transcriptomic level.
Similar content being viewed by others
Introduction
Inflammatory bowel disease (IBD) includes both Crohn's disease (CD) and ulcerative colitis (UC) and results from improper inflammatory response to microbiota in a host with genetic susceptibility1. With tumor necrosis factor alpha (TNF-α) being the key cytokine in inflammation, many anti-TNF therapies emerged and dominated in the treatment of CD2,3. On one hand, disease management was significantly improved with anti-TNF agents, such as adalimumab (ADA) and infliximab (IFX)4,5, but on the other hand, 30% of patients do not respond to the anti-TNF treatment and of those who initially benefit, up to 40% lose their response later2,4,6,7. Moreover, unnecessary continuation of the anti-TNF therapy may severely impede patients' quality of life and impose adverse effect risk without clinical justification with emerging of an additional challenge of loss of response management8,9. Thus, identification of anti-TNF therapy response predictors is of pivotal importance and is posing an additionally challenge in the last decade. Prediction of response is also needed as we now have other therapies with different targets available10. Several studies have focused on genetic background and gene expression11,12,13,14,15,16,17 as potential biomarkers for anti-TNF treatment response. Currently the genomics markers of anti-TNF response in CD are not reaching significant thresholds and practically no reproducibility between genetic and expression markers exists, as identified by the first integration of genomic and expression data prior to anti-TNF treatment in a systematic review18. Prediction profiles have been mostly identified using epithelial gene expression data for which colonoscopy and biopsy sampling are needed16,17. However, obtaining colon biopsies is a highly invasive procedure. Studies which would focus on non-invasive genetic expression biomarkers, such as gene expression in peripheral blood mononuclear cells (PBMC) for anti-TNF response identification are scarce and non-homogenous in terms of anti-TNF therapy and disease classification.
Therefore, the aim of the present study is to integrate transcriptomic and genomic data obtained from blood of CD patients, who were naïve with respect to the treatment with biologicals, using our and previously reported peripheral blood derived transcriptomic data and using a modified contemporary combination of omics approach as described previously19. For that, we integrated our omics data based on 46 genes identified in CD and rheumatoid arthritis (RA) cohorts, which appeared the most significant to differentiate responders from non-responders to anti-TNF therapy (Table 1)20. We also included four genes that correlated with changes in endoscopic activity in UC patients with anti-TNF therapy and three differentially expressed genes between responding and non-responding IBD patients to anti-TNF therapy as identified by a cell-centered meta-analysis (Table 1)21,22. Additionally we included 12 independently confirmed and previously integrated genomic and expression (RNA and protein) markers of anti-TNF therapy response in CD patients (Table 1)18. The investigation gene panel built for the present study was altogether based on 65 previously suggested candidate genes in 2126 individuals with IBD and 34 individuals with RA, and treated with anti-TNF therapy.
Results
RNA-seq analysis
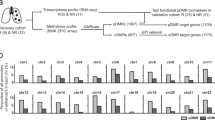
Using our RNA sequencing data we performed an expression study using previously suggested candidate genes discriminating anti-TNF response in CD patients. For that, we used two different approaches. In first approach, the analysis was adjusted to deconvoluted fractions of PBMCs and has identified 11 differentially expressed genes out of which MMD, ELVOL7 and BMP6 remained significantly down-regulated in responders relative to non-responders after correction (Table 2, Fig. 1). In the second approach, analysis was carried out unadjusted in order to obtain PBMC panel. The analysis has confirmed 10 differentially expressed genes out of which 4 (GPR84, EPSTI1, IFI6, MX1) remained significantly up-regulated after correction for multiple comparison in responders relative to non-responders (Table 2, Fig. 1).
RNA-seq volcano and smear plots of RNA-seq analyses. RNA-seq analysis. The plots were constructed using ggplot2 R package23.
Integration to genomics
Using an integrative transcriptomic-genomic approach we analyzed single nucleotide variants ranging ± 100 kb from previously identified differentially expressed genes in 84 patients with CD. Association analysis has identified 5 statistically significant variants near gene MMD (Fig. 2A) and 2 statistically significant variants in ELOVL7 gene region (Fig. 2B). For single nucleotide polymorphisms (SNPs) rs1465352, rs4422035, rs9892429, rs9893820 and rs11656799 near gene MMD the results have shown significant higher alternative allele frequency in non-responders in comparison to responders (Table 3). For the most significant rs4422035 the analysis has shown higher alternative allele frequencies in non-responders (Allele C: 0.69) in comparison to responders (Allele C: 0.45) (p = 0.018; OR 0.22; CI 0.09–0.55). Additional LD assessment has shown that that rs4422035 and rs1465352 are in linkage disequilibrium (D′: 1.0; r2: 0.87; p < 10–4). Identified rs9892429, rs9893820 and rs11656799 have shown higher linkage disequilibrium with rs1465352 (D′: 1.0; r2: 0.96; p < 10–4) as in comparison to most significant rs4422035, which is also evident from the Table 3. Similar effect was observed also for variants identified in ELOVL7 gene region. Both identified variants have shown the same trend of alternative allele frequencies (Table 3). For the most significant rs78620886 the analysis has shown higher alternative allele frequencies in non-responders (Allele A: 0.26) in comparison to responders (Allele A: 0.03) (p = 0.021; OR 0.05; CI 0.01–0.27). Additional LD analysis has also shown that rs78620886 is in linkage disequilibrium with rs9291695 (D′: 1.0; r2: 0.95; p < 10–4).
Regional Manhattan plots for MMD and ELOVL7 ± 100 kb gene region. (A) MMD; (B) ELOVL7. Regional Manhattan plots were constructed using LocusZoom24.
Functional analysis of identified variants
Identified variants were further assessed for possible functional effects. All seven variants correspond to locations on non-coding DNA region. SNPs rs1465352 and rs4422035 near gene MMD were listed at H3K9ac_Pro histone acetylation mark in monocytes. Also the ELOVL7 intronic rs78620886 was listed at the H3K9ac_Pro mark in primary T helper naive cells derived from peripheral blood (Table 4). Subsequently, eQTLs were also assessed (Table 4). For rs1465352 and rs11656766 significant eQTLs with MMD were listed in monocytes. For rs4422035, rs9892429 and rs9893820 no eQTL data was available in monocytes. For rs9291695 and rs78620886, significant eQTLs with ELOVL7 were listed in EBV transformed lymphocytes. Additionally, genotyping results from rs1465352, rs4422035 and rs78620886 were used as variables for prediction value analysis using random forest classification and ROC analysis. Obtained results further confirmed the association of rs1465352, rs4422035 and rs78620886 in adalimumab response (AUC 0.833; CI 0.740–0.926) with sensitivity of 0.84, specificity of 0.72 and Youden index of 0.56 at the best cut-off point (Fig. 3).
ROC curve of random forest classification of rs1465352, rs4422035 and rs78620886 genotypes for adalimumab response prediction. The ROC curve plot was constructed using ggplot2 R package23.
Mendelian randomization analysis
An additional Inverse-variance random-effect Mendelian Randomization (MR) analysis was performed. First, variants ranging ± 100 kb from MMD and ELOVL7 were included into conditional regressions in order to detect independent signals conditioned to the identified most significant SNPs. Conditioning results have identified statistically significant rs12943795 besides rs4422035 for MMD, and rs182724486 and rs149880548 besides rs78620886 for ELOVL7. Statistically significant conditioned independent variants were further assessed for eQTLs in corresponding tissues/cells in which chromatin state assignment was listed in HaploReg and were included into subsequent MR analysis. MR analysis has proven the association of ELOVL7 with response to adalimumab (p = 0.046; SE 2.812; CI95 11.13 to − 0.11). For MMD no MR analysis was performed due to unavailable eQTLs for independent rs12943795 and rs4422035 SNPs in corresponding tissues/cells.
Validation of genes using RT-qPCR
In order to assess and validate the findings of RNA-seq analysis and subsequent genomic variants, RT-qPCR was performed. First, reference genes were checked for stability between responders and non-responders. Both genes, ACTB (p = 0.768) and B2M (p = 0.782) proved to be stably expressed in both groups. Subsequently gene expression of MMD and ELOVL7 was measured in PBMCs. Both genes were down-regulated in responders, but statistical significance was observed only at ELOVL7, which was 1.43-fold down-regulated in responders relative to non-responders (p = 0.016; β − 0.535; SE 0.214) (Fig. 4B). MMD was 1.10-fold down-regulated in responders relative to non-responders (p = 0.498; β − 0.141; SE 0.656) (Fig. 4A). Additionally eQTLs in PBMCs were assessed for SNPs where chromatin state assignment was listed. No statistically significant eQTLs were observed for rs1465352 (p = 0.564; β 0.002; SE 0.004) (Fig. 4C), rs4422035 (p = 0.752; β 0.001; SE 0.004) (Fig. 4D) and rs78620886 (p = 0.448; β 0.003; SE 0.004) (Fig. 4E). In order to check the interplay of MMD and ELOVL7 a correlation analysis of expression was performed. Correlation analysis revealed a strong correlation between genes (p = 2.57 × 10–13; ρ = 0.799) (Fig. 4F).
Validation of results using RT-qPCR, eQTL analysis and correlation estimation. (A) MMD relative expression; (B) ELOVL7 relative expression; (C) eQTL for rs1465352 and MMD; (D) eQTL for rs4422035 and MMD; (E) eQTL for rs78620886 and ELOVL7; (F) Correlation between MMD and ELOVL7 expression. Black lines represent median expression. Blue line represents correlation regression curve with standard error. The plots were constructed using ggplot2 R package23.
Discussion
Anti-TNF therapy is of pivotal importance in patients with CD and genomic biomarkers that would tailor personalized treatment are currently needed, in particular as new biologicals for CD treatment have recently emerged. In the recent systematic review of anti-TNF genomic biomarkers in CD, it was noted that almost no candidate gene could be confirmed in an independent association study and no overlap was detected among genes identified with association and gene expression studies18. In the present study, we have adopted a new approach19 for the integration of PBMCs derived transcriptomic and genomic data. To our knowledge, this is the first time deconvolution was applied in order to account for PBMC composition in anti-TNF response transcriptomics, which identified two novel genetic loci on chromosomes 17 near gene MMD and 5 in gene ELOVL7, associated with anti-TNF response in CD patients.
For that, we included patients with CD, who were naïve with respect to the treatment with biologicals and who were subsequently treated with ADA and investigated PBMC derived transcriptomic-genomic data integration with targeting on previously identified gene expression panels for differentiation of responders from non-responders to anti-TNF therapy. Additionally, we investigated independently confirmed and previously integrated genomic and expression (RNA and protein) markers of anti-TNF therapy response in CD patients. Thus, the targeted expression panel consisted altogether of 65 genes, which were previously identified in 2126 individuals with IBD and 34 individuals with RA as potential predictors for different anti-TNF therapy (ADA, IFX, golimumab) responses in peripheral blood of CD, UC, unclassified IBD or RA patients18,20,21,22.
First, using RNA-seq and using 3 responders and 3 non-responders to ADA we confirmed differentially expressed genes from previously identified gene panel. The model fitting for estimation of differentially expressed genes was carried out in two different ways—adjusted to deconvolution results and not adjusted in order to obtain PBMC expression picture. The results have shown that in deconvoluted analysis MMD, ELOVL7 and BMP6 remained significantly differentially expressed after correction for multiple comparisons. All three genes were down-regulated in responders relative to non-responders (up-regulated in non-responders relative to responders). Using unadjusted model fitting we observed the confirmation of GPR84, EPSTI1, IFI6, MX1, which remained significantly differentially expressed after the application of multiple comparison correction. All four were found to be up-regulated in responders relative to non-responders (down-regulated in non-responders relative to responders). Subsequently, confirmed genes were further analyzed using integration with genomic data obtained from 84 CD patients, who were naïve with respect to the treatment with biologicals, using a genotype microarray and using an association analysis with 12 week response as an outcome variable. Imputed high quality single nucleotide variants were analyzed using a margin of ± 100 kb from confirmed genes in RNA-seq analysis in order to include variants in regulatory gene regions. The results have identified two significant signals, one near gene MMD and one in ELOVL7 gene region after application of Bonferroni-like correction using empirical autocorrelation estimation, but no statistically significant signals were observed in other 5 genes. Both MMD and ELOVL7 were identified in deconvoluted approach. Thus, obtained results are pointing to increased robustness of analysis when using deconvolution of PBMCs. As previously shown using deconvolution-meta-analysis paradigm, adjustment to estimation of cell proportions can capture differential expression otherwise masked due to cellular composition variation22. Expression of MMD (Monocyte to macrophage differentiation-associated) gene can be detected in all tissues and its’ up-regulation is observed upon monocyte differentiation, which modulates the production of TNF-α and nitrous oxide (NO)25. TNF was previously identified among key players of the Schmitt et al. proposed model of anti-TNF non-response26 and was identified as a sole key player in the baseline response prediction screening18. It was also previously suggested that there is an association between the loss of anti-TNF response and ROS response27, which is in concordance with our findings as up-regulation of MMD gene modulates also the production of NO. ELOVL7 (ELOVL Fatty acid elongase 7) gene is a member of 7 ELOVL isozymes which catalyze the first step of very long-chain fatty acid elongation cycle28. In the context of IBD, ELOVL7 was previously associated only with protective association for hidradenitis suppurativa29. In aforementioned regions, the analysis identified SNPs rs1465352, rs4422035, rs9892429, rs9893820 and rs11656766 located downstream of MMD and SNPs rs9291695 and rs78620886 located in ELOVL7 intronic regions. OR calculated for alternative allele for all SNPs indicates higher probability for non-responding to ADA therapy. To the best of our knowledge this is the first time that the aforementioned SNPs are associated with response to anti-TNF therapy in CD. Furthermore, functional analysis for MMD rs1465352 and rs4422035, and ELOVL7 rs78620886 has shown that these SNPs are listed in HaploReg v4.1 at H3K9ac_Pro histone modification epigenetic mark in monocytes and lymphocytes, respectively. Histone modifications are implicated in influencing gene expression by establishing global chromatin environments and H3K9ac (Histone 3 Lysine 9 acetylation) histone modification is enriched in promoter regions containing critical regulatory elements necessary for transcription30. H3K9 is a residue that can be acetylated or methylated and H3K9ac was shown to be dramatically reduced during chromosome condensation31. Moreover, H3K9ac is associated with active promoters and is considered as a hallmark of active transcriptions32. As MMD and ELOVL7 were both up-regulated in non-responders to ADA, we hypothesize that locations of rs1465352, rs4422035 and rs78620886 might play a significant role in histone modifications in terms of H3K9 residue acetylation and up-regulation of gene expression. Using ENSEMBL33 and GTExportal database34 statistically significant eQTLs with MMD and ELOVL7 were also listed. Additionally, the implication of aforementioned SNPs in ADA response was also confirmed using random forest machine learning algorithm and ROC analysis, which has additionally confirmed the association of these three SNPs with ADA response in patients with CD. In order to fully address the implication of both genes in ADA response, a Mendelian randomization integrating eQTL and association analysis data was performed. Gene ELOVL7 was subsequently again confirmed as a significant candidate, however no analysis was performed for MMD due to unavailability of eQTL data for selected variants in corresponding tissues/cells in used databases33,34. Furthermore, monocyte to macrophage differentiation was found to go along with profound changes in lipid-related transcriptome, leading to an induction of fatty-acid elongation35, which can elucidate the interplay of MMD and ELOVL7 up-regulation in ADA non-responders. These findings are further supported by previously shown genetically influenced abnormalities in fatty acid profiles in IBD36. The main limitation of the present study is the limited sample size in transcriptome analysis. Nevertheless, we believe that using trimmed mean of M values method (TMM) normalization implemented in edgeR and the variance modeling at the observational level (VOOM) method implemented in the limma package before applying empirical Bayes over a linear model, made our analysis stringent enough to provide reliable results. The aforementioned concern was also addressed using RT-qPCR analysis for estimation of MMD and ELOVL7 gene expression in PBMCs in a larger cohort in order to validate the findings. However, MMD and ELOVL7 were identified using an approach with deconvolution of PBMCs, thus differences between responders and non-responders may not have been fully revealed using RT-qPCR. Obtained results have shown that both, MMD and ELOVL7 tend to be up-regulated in non-responders relative to responders, but only ELOVL7 reached statistical significance, which is consistent with RNA-seq findings. Furthermore, for SNPs where chromatin state assignment was listed, no trend of association with MMD and ELOVL7 was observed in PBMCs, which is also consistent with reported eQTLs in whole blood. Moreover, using RT-qPCR we observed a strong correlation between MMD and ELOVL7, which further supports the interplay between these two genes and serves as additional supporting evidence of monocyte to macrophage differentiation and changes in lipid-related transcriptome. In addition, based on our results functional studies are warranted and could further unravel the interplay between MMD, ELOVL7 and H3K9ac in response to anti-TNF therapy. We also acknowledge that the homogenous group of enrolled IBD classified CD patients and using only one anti-TNF agent represents the strength of our study design.
In summary, the present study integrated transcriptomic and genomic data obtained from CD patients’ PBMCs and identified two novel genetic loci, on chromosomes 17 and 5, associated with anti TNF response in CD patients and suggested MMD and ELOVL7 as best candidates for involvement in anti-TNF response. Moreover, the present study revealed that the implication and regulation of MMD and ELOVL7 gene regions in ADA response may be a part of a complex interplay extending from genetic to epigenetic and to transcriptomic level. This knowledge could be further translated into new clinical non-invasive baseline biomarkers for adalimumab response in patients with CD.
Methods
All methods were carried out in accordance with relevant guidelines and regulations.
Enrolled subjects
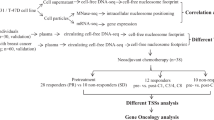
We included 84 Slovenian patients with CD who fulfilled the criteria for anti-TNF-α drug ADA (Humira, Abbott Laboratories, IL, USA), and who were naïve with respect to the treatment with biologicals. All enrolled subjects were of Slovenian (Caucasian—Central Europe) ethnicity. Baseline demographics of the patients are shown in Table 5. Inclusion criteria were refractoriness to corticosteroids, adverse effects to corticosteroids and lost response or intolerance to IFX as described previously12. Exclusion criteria were defined as presence of CD complications as stenosis, abscesses, total colectomy, history of murine proteins allergy, active tuberculosis or a serious infection in the last 3 months, pregnancy or lactation and malignancy37. First, a loading dose of 160 mg of ADA was administered, followed by 80 mg after 14 days and maintenance dose of 40 mg every other week. Azathioprine, 5-aminosalycylates, corticosteroids or antibiotic concomitant treatment was allowed if the dosage was stable in the last 3 months. The present study was evaluated and approved by Slovenian National Committee for Medical Ethics (KME 80/10/07, 21p/12/07). Written informed consent was obtained from all enrolled subjects. In case of minors, written informed consent was obtained from parents and/or legal guardians. Response to treatment was measured using IBD questionnaire (IBDQ) after 12 weeks of treatment. Response was defined as an increase in score > 22 points in comparison of pre-treatment score or total score > 170 points38,39.
Extraction of nucleic acids
DNA and RNA were extracted from peripheral blood mononuclear cells (PBMC) using TRI-reagent (Merck, Darmstadt, Germany) according to manufacturer’s instructions. Purity and concentration of nucleic acids was determined using Synergy 2 spectrophotometer (Biotek, Winooski, VT, USA). Integrity of RNA was checked using agarose gel electrophoresis and 2100 Bioanalyzer Instrument (Agilent, Santa Clara, CA, USA) with RNA 6000 Nanochip.
RNA-seq analysis
For RNA-seq analysis, 3 responders and 3 non-responders were selected. These patients were selected for RNA-seq based on available data for continuously measured treatment response, which was recorded at weeks 4, 12, 20 and 30. The selected 6 patients qualified as 3 were consistent responders and 3 consistent non-responders during all check point measurements. RNA RIN number was > 9.0 and 28S/18S ratio between 1.5 and 2.0 for all 6 samples. Both, lncRNA and mRNA 100 bp paired-end libraries were constructed using MGIEasy rRNA Depletion Kit (MGITech, Shenzen, China) and MGIEasy RNA Library Prep Set (MGITech) and using the BGISEQ-500 instrument (BGI, Hong Kong) at BGI facilities (BGI). Data analysis was performed using R 4.0.2 environment (R Core Team 2020, Vienna, Austria). Raw .fastq files were first assessed for quality using FastQC 0.11.9 software40. Technical sequences and sequencing adapters were trimmed using Trimmomatic 0.39 tool41. Paired-end reads were aligned to the hg19 reference genome using Rsubread 2.2.4 R package42,43. Mapped reads were counted and assigned to genomic features using featureCounts44 with requirement of both ends to be mapped. Counts per million (CPMs) were calculated using edgeR 3.30.3 R package45. Low expressed genes were filtered out based on CPMs corresponding to read counts of 10 and retained genes were normalized using the trimmed mean of M values method (TMM)46. Subsequently, mean–variance modeling at the observational level transformation (VOOM) was applied47. Differential expression of responders relative to non-responders was determined using linear models and empirical bayes implemented in limma 3.44.3 R package48 using two approaches. In first approach, the linear model was adjusted based on the deconvoluted peripheral blood mononuclear cell composition data. Deconvolution was used to obtain an estimation of the abundances of member cell types in a mixed cell population, using gene expression data in terms of proportions of different white cell subtypes in PBMCs49. Using raw counts, transcripts per million (TPM) were calculated, zero values were removed and data was deconvoluted using CIBERSORT49 and LM2249 signature matrix. Sample composition in terms of proportions of lymphocytes, monocytes/macrophages and neutrophils was used as a covariate in the model. Zero estimated cell proportions were excluded from analyses. In the second approach, the linear model was left unadjusted in order to obtain PBMC gene profile picture. Differential expression was considered for genes with adjusted p-value < 0.05.
Association analysis
Samples obtained from 84 enrolled individuals were genotyped using Infinium Global Screening Array (GSA_24v1) (Illumina, San Diego, California, USA). Quality control of the genotyped data was performed as described previously50. Further steps of analysis were based on an integrative transcriptomic-genomic approach19. Subsequently, imputation was performed using Michigan Imputation Server Minimac3 genotype imputation algorithm and using HRC r1.1 2016 reference panel and SHAPEIT v2.r790 phasing51. Association between responders at week 12 vs. non-responders at week 12 was tested using binary logistic Wald test implemented in EPACTS 3.2.652. Association analysis was adjusted to age at diagnosis, sex, azathioprine use, use of aminosalycylates, use of corticosteroids and first four principal components, which were calculated using PLINK 1.9 software53,54. Appropriate number of principal components was determined beforehand using gap v1.2.2 R package. Retained were only variants with imputation score (Rsq) > 0.3. Variants ± 100 kb from previously identified differentially expressed genes were further analyzed. Bonferroni-like correction was applied based on empirical autocorrelation determined using coda R package55. Statistically significant signal was considered for SNPs with adjusted p-value < 0.05. Linkage disequilibrium was calculated using LDlink software56.
In silico functional analysis, prediction value estimation and visualization
Functional analyses were done using HaploReg v4.157, GTExPortal34 and ENSEMBL33. Regional Manhattan plots were constructed using LocusZoom24. At SNPs, where chromatin state assignment was listed, predictive value of genotypes was further assessed using randomForest 4.6-14 R package 58 and Receiver Operating Characteristics analysis using pROC R package59. All other plots were constructed using ggplot2 R package23.
Mendelian randomization analysis
Data was further analyzed using Mendelian randomization integration of eQTL and association analysis data. Previously retained variants ± 100 kb from MMD and ELOVL7 gene regions were further analyzed using GCTA-COJO conditional analysis60. GCTA-COJO conditional analysis was performed conditional on most significant SNP identified in previous association analysis. Reference dataset for GCTA-COJO was built based on previously imputed data. Variants with Rsq < 0.3 were removed from the reference dataset prior to analysis. Condition SNP and SNPs, which remained significant after conditioning, were included in subsequent Mendelian randomization analysis. Mendelian randomization was performed using MendelianRandomization v0.5.0 R package and inverse-variance weighted random-effect method61. Variant-gene association (eQTL) obtained from GTExportal34 and ENSEMBL33 in corresponding tissues/cells was included as exposure and variant-outcome association as outcome for MR analysis. If no eQTL data was available in corresponding tissues/cells, the variant was excluded from MR analysis.
Validation of genes using RT-qPCR
A total of 1 µg of mRNA with RIN > 8 was obtained from 55 (21 non-responders and 34 responders) out of 84 enrolled individuals and was transcribed into cDNA using high capacity cDNA reverse transcription kit (Thermo Fisher, Waltham, MA, USA). mRNA nucleotide sequences of target genes MMD and ELOVL7 were retrieved from NCBI Nucleotide database (www.ncbi.nlm.nih.gov/nuccore/). Primers for reference genes ACTB and B2M were obtained from previous study17. Isoform non-specific primers were hand-picked and designed using IDT OligoAnalyzer Tool (eu.idtdna.com/calc/analyzer). Primer sequences and accession numbers are summarized in Table 6. All primers were synthesized by Sigma (Merck, Darmstadt, Germany). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) gene expression assay was carried out using Lightcycler 480 SYBR Green I Master Mix and Lightcycler 480 real time thermocycler (Roche, Basel, Switzerland) according to manufacturer’s instructions. 2 µL of 20-fold diluted cDNA (2.5 ng/µL) was used as a template. Primer efficiency was > 90% for all primer pairs. Melting curves for each sample were analyzed after each run in order to confirm specificity of amplification. Raw Ct values were obtained from three run-independent technical replicates for each sample. Normalization of raw data was performed using geometric averaging of reference genes and relative expression was calculated using 2−ΔΔCt method62. Statistics was performed using linear 2−ΔCt calculation and linear regression was adjusted to same covariates as aforementioned in association analysis. Stability of reference genes was statistically assessed using relative 2−ΔCt calculation and Mann–Whitney U-Test. eQTLs based on the study data were estimated for SNPs where chromatin state assignment was listed. Estimation was performed using linear regression. Violation of linear regression assumption was considered if variance inflation factor > 10 and condition index > 30. Correlation between gene expressions was assessed using Spearman’s rank correlation.
References
Abraham, C. & Cho, J. H. Inflammatory bowel disease. N. Engl. J. Med. 361, 2066–2078 (2009).
Danese, S., Fiorino, G. & Reinisch, W. Review article: Causative factors and the clinical management of patients with Crohn’s disease who lose response to anti-TNF-alpha therapy. Aliment Pharmacol. Ther. 34, 1–10 (2011).
Van Deventer, S. J. Tumour necrosis factor and Crohn’s disease. Gut 40, 443–448 (1997).
Colombel, J. F. et al. Adalimumab for maintenance of clinical response and remission in patients with Crohn’s disease: The CHARM trial. Gastroenterology 132, 52–65 (2007).
Hanauer, S. B. et al. Maintenance infliximab for Crohn’s disease: The ACCENT I randomised trial. Lancet 359, 1541–1549 (2002).
Sandborn, W. J. et al. Adalimumab for maintenance treatment of Crohn’s disease: Results of the CLASSIC II trial. Gut 56, 1232–1239 (2007).
Sands, B. E. et al. Infliximab maintenance therapy for fistulizing Crohn’s disease. N. Engl. J. Med. 350, 876–885 (2004).
Ben-Horin, S., Kopylov, U. & Chowers, Y. Optimizing anti-TNF treatments in inflammatory bowel disease. Autoimmun. Rev. 13, 24–30 (2014).
Restellini, S. et al. Therapeutic drug monitoring guides the management of Crohn’s patients with secondary loss of response to adalimumab. Inflamm. Bowel Dis. 24, 1531–1538 (2018).
Schoultz, I. & Keita, A. V. Cellular and molecular therapeutic targets in inflammatory bowel disease-focusing on intestinal barrier function. Cells 8, 193 (2019).
Dezelak, M., Repnik, K., Koder, S., Ferkolj, I. & Potocnik, U. A prospective pharmacogenomic study of Crohn’s disease patients during routine therapy with anti-TNF-alpha drug adalimumab: Contribution of ATG5, NFKB1, and CRP genes to pharmacodynamic variability. OMICS 20, 296–309 (2016).
Koder, S. et al. Genetic polymorphism in ATG16L1 gene influences the response to adalimumab in Crohn’s disease patients. Pharmacogenomics 16, 191–204 (2015).
Repnik, K., Koder, S., Skok, P., Ferkolj, I. & Potocnik, U. Transferrin level before treatment and genetic polymorphism in HFE gene as predictive markers for response to adalimumab in Crohn’s disease patients. Biochem. Genet. 54, 476–486 (2016).
Barber, G. E. et al. Genetic markers predict primary non-response and durable response to anti-TNF biologic therapies in Crohn’s disease. Am. J. Gastroenterol. 111, 1816–1822 (2016).
Yoon, S. M. et al. Colonic phenotypes are associated with poorer response to anti-TNF therapies in patients with IBD. Inflamm. Bowel Dis. 23, 1382–1393 (2017).
Arijs, I. et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS ONE 4, e7984 (2009).
Gorenjak, M. et al. Genetic prediction profile for adalimumab response in Slovenian Crohn’s disease patients. Z. Gastroenterol. 57, 1218–1225 (2019).
Gole, B. & Potocnik, U. Pre-treatment biomarkers of anti-tumour necrosis factor therapy response in Crohn’s disease—A systematic review and gene ontology analysis. Cells 8, 515 (2019).
Hernandez-Pacheco, N. et al. Combined analysis of transcriptomic and genetic data for the identification of loci involved in glucocorticosteroid response in asthma. Allergy 00, 1-6 (2020).
Mesko, B. et al. Peripheral blood derived gene panels predict response to infliximab in rheumatoid arthritis and Crohn’s disease. Genome Med. 5, 59 (2013).
Planell, N. et al. Usefulness of transcriptional blood biomarkers as a non-invasive surrogate marker of mucosal healing and endoscopic response in ulcerative colitis. J. Crohns Colitis 11, 1335–1346 (2017).
Gaujoux, R. et al. Cell-centred meta-analysis reveals baseline predictors of anti-TNFalpha non-response in biopsy and blood of patients with IBD. Gut 68, 604–614 (2019).
Wickham, H. et al. ggplot2: Elegant graphics for data analysis (Springer, 2016).
Pruim, R. J. et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Liu, Q. et al. Monocyte to macrophage differentiation-associated (MMD) positively regulates ERK and Akt activation and TNF-alpha and NO production in macrophages. Mol. Biol. Rep. 39, 5643–5650 (2012).
Schmitt, H. et al. Expansion of IL-23 receptor bearing TNFR2+ T cells is associated with molecular resistance to anti-TNF therapy in Crohn’s disease. Gut 68, 814–828 (2019).
Luther, J. et al. Loss of response to anti-tumor necrosis factor alpha therapy in Crohn’s disease is not associated with emergence of novel inflammatory pathways. Dig. Dis. Sci. 63, 738–745 (2018).
Leonard, A. E., Pereira, S. L., Sprecher, H. & Huang, Y. S. Elongation of long-chain fatty acids. Prog. Lipid Res. 43, 36–54 (2004).
Janse, I. C. et al. Identification of clinical and genetic parameters associated with hidradenitis suppurativa in inflammatory bowel disease. Inflamm. Bowel Dis. 22, 106–113 (2016).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Park, J. A. et al. Deacetylation and methylation at histone H3 lysine 9 (H3K9) coordinate chromosome condensation during cell cycle progression. Mol. Cells 31, 343–349 (2011).
Karmodiya, K., Krebs, A. R., Oulad-Abdelghani, M., Kimura, H. & Tora, L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genom. 13, 424 (2012).
Hunt, S. E. et al. Ensembl variation resources. Database (Oxford) 2018 (2018).
Consortium, G. Erratum: Genetic effects on gene expression across human tissues. Nature 553, 530 (2018).
Wallner, S. et al. Monocyte to macrophage differentiation goes along with modulation of the plasmalogen pattern through transcriptional regulation. PLoS ONE 9, e94102 (2014).
Jezernik, G. & Potocnik, U. Comprehensive genetic study of fatty acids helps explain the role of noncoding inflammatory bowel disease associated SNPs and fatty acid metabolism in disease pathogenesis. Prostaglandins Leukot. Essent. Fatty Acids 130, 1–10 (2018).
Panaccione, R. et al. Efficacy and safety of adalimumab in Canadian patients with moderate to severe Crohn’s disease: Results of the Adalimumab in Canadian SubjeCts with ModErate to Severe Crohn’sDiseaSe (ACCESS) trial. Can. J. Gastroenterol. 25, 419–425 (2011).
Hlavaty, T. et al. Evaluation of short-term responsiveness and cutoff values of inflammatory bowel disease questionnaire in Crohn’s disease. Inflamm. Bowel Dis. 12, 199–204 (2006).
Guyatt, G. et al. A new measure of health status for clinical trials in inflammatory bowel disease. Gastroenterology 96, 804–810 (1989).
Andrews, S. FastQC: A quality control tool for high throughput sequence data (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Liao, Y., Smyth, G. K. & Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019).
Liao, Y., Smyth, G. K. & Shi, W. TheSubread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Anderson, C. A. et al. Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573 (2010).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Kang, H. M. EPACTS: Efficient and parallelizable association container toolbox. (2016).
Chang, C. C. et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Plummer, M., Best, N., Cowles, K. & Vines, K. CODA: Convergence diagnosis and output analysis for MCMC. R News 6, 7–11 (2006).
Machiela, M. J. & Chanock, S. J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015).
Ward, L. D. & Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930-934 (2012).
Liaw, A. & Wiener, M. Classification and regression by randomForest. R News 2, 18–22 (2002).
Robin, X. et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Yavorska, O. O. & Burgess, S. MendelianRandomization: An R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 46, 1734–1739 (2017).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta DeltaC(T)) Method. Methods 25, 402–408 (2001).
Acknowledgements
The authors would like to thank the patients for participation in this study. The authors would like to thank Ms. Dr. Natalia Hernandez-Pacheco, MSc, PhD and Ms. Dr. Maria Pino-Yanes, PhD for advices and support. The authors would like to thank Dr. Boris Gole, PhD for a careful review and recommendations.
Funding
The authors acknowledge the project (Molecular genetic biomarkers and mechanisms of non-response to biological treatment with anti-TNF in patients with IBD, ID J3-9258) was financially supported by the Slovenian Research Agency. The Slovenian Research Agency had no role in the design, execution, interpretation, or writing of the study. M.Z. was founded by Ministry of Education, Science and Sport (C3330-19-952026).
Author information
Authors and Affiliations
Contributions
M.G. and U.P. were involved in the conception and design of the study. M.G., M.Z., G.J. and P.S. were involved in the data acquisition. M.G., M.Z. and G.J. contributed to the analysis of the data. M.G., P.S. and U.P. contributed to the interpretation of the results. All authors drafted the article. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gorenjak, M., Zupin, M., Jezernik, G. et al. Omics data integration identifies ELOVL7 and MMD gene regions as novel loci for adalimumab response in patients with Crohn’s disease. Sci Rep 11, 5449 (2021). https://doi.org/10.1038/s41598-021-84909-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-84909-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.