« Prev Next »
The proteome is neither as uniform nor as static as the genome. Whereas the genetic code is made of four nucleotides and the sequence of these nucleotides is identical in every cell of an organism, proteins are built from 20 different amino acids, and post-translational modification adds other chemical constituents to these molecules, including sugars, fats, phosphates, and even other proteins! In addition, proteins come in different isoforms, are churned through metabolic and degradative pathways, are alternatively spliced, and often link with one another to form complexes made up of multiple proteins. Moreover, the set of proteins produced by a cell varies depending on cell type, cell shape, cell function, what tissue the cell resides in, and what signals the cell receives from its environment, not to mention what developmental stage the cell is in.
Benefits of Proteomics
Proteomics doesn't only reveal information about life's complexity, however; it also provides insight into the vibrancy of cells and their preparedness to react. Cells and tissues respond to signals and changes in their environment, and changes in the proteome must mirror that. In fact, early changes in the health of a tissue may be detectable by changes at the proteomic level. Researchers are just beginning to take advantage of measurable changes in protein profiles to assess disease; for example, assaying serum proteins using a chip-based mass spectrometry system reveals a difference in protein profiles between men who have benignly enlarge prostates and those who have prostate cancer (Adam et al., 2002). Moreover, the difference in profiles is robust enough to use as a predictive diagnostic tool.
Studying the Proteome
The first complete eukaryotic genomic sequence was of the yeast Saccharomyces cerevisiae (Goffeau et al., 1996). Prior to sequencing, a rich knowledge of yeast biology had already been acquired through decades of hypothesis-driven research with this model organism. Today, even more genomics data and information resources are available to scientists, including the Yeast Protein Database and the Saccharomyces Genome Database. An ambitious study sought to integrate these genomic and proteomic data after experimental manipulation of a well-studied metabolic pathway in yeast, the galactose utilization pathway. A mathematical model was created and used to predict previously unknown interactions within the pathway and with other cellular processes. Some of these predictions were then verified experimentally (Ideker et al., 2001).
Systems biology approaches will continue to detect connections between broad cellular functions and pathways that were neither apparent nor predictable despite decades of biochemical and genetic analysis of the biological system in question. For example, one study looked at quantitative protein profiling in cells with and without the oncogene Myc, one of the most frequently altered genes in human cancer (Shiio et al., 2002). Here, the researchers noted differences in the adhesion molecules associated with these cells—differences that may underlie the morphological changes that lead to unchecked proliferation in cancer. This finding represents how proteomics can enrich our understanding of the interdependence of cellular processes.
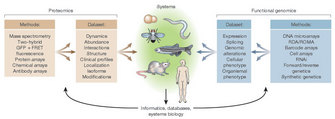
Currently, researchers' ability to collect large proteomic data sets is greater than their ability to interpret or integrate that information. Thus, the need for bioinformatics algorithms and software tools will likely remain high for some time to come. While improvements in proteomic technologies will likely accelerate research involving single-celled organisms, the additional layers of complexity and organization in multicellular organisms will necessitate grander conceptual schemes, such as those associated with systems biology (Figure 3). Beyond relating genes to transcripts, transcripts to proteins, or proteins to functions, systems biology seeks to integrate all of these layers to achieve a fuller understanding of normal function, disease, and development.
References and Recommended Reading
Banks, R. E. Proteomics: New perspectives, new biomedical opportunities. Lancet 356, 1749–1756 (2000) doi:10.1016/S0140-6736(00)03214-1
Brett, D., et al. Alternative splicing and genome complexity. Nature Genetics 30, 23–30 (2001) doi:10.1038/ng803 (link to article)
Goffeau, A., et al. Life with 6000 genes. Science 274, 546–567 (1996)
Ideker, T., et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292, 929–934 (2001)
Jensen, O. N. Interpreting the protein language using proteomics. Nature Reviews Molecular Cell Biology 6, 391–403 (2006) doi:10.1038/nrm1939 (link to article)
Kalia, A., & Gupta, R. P. Proteomics: A paradigm shift. Critical Review of Biotechnology 25, 173–198 (2005)
O'Farrell, P. High resolution two-dimensional electrophoresis of proteins. Journal of Biological Chemistry 250, 4007–4021 (1975)
Ozbal, C. C., et al. High throughput screening via mass spectrometry: A case study using acetylcholinesterase. Assay Drug Development Technologies 2, 373–381 (2004)
Schmucker, D., et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671–684 (2000)
Shiio, Y., et al. Quantitative proteomic analysis of Myc oncoprotein function. EMBO Journal 21, 5088–5096 (2002) doi:10.1093/emboj/cdf525
Stumpf, M. P. H., et al. Estimating the size of the human interactome. Proceedings of the National Academy of Sciences 105, 6959–6964 (2008) doi:10.1073/pnas.0708078105
Tyers, M., & Mann, M. From genomics to proteomics. Nature 422, 193–197 (2003) doi:10.1038/nature01510 (link to article)
Ullrich, B., Ushkaryov, Y. A., & Südhof, T. C. Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron 14, 497–507 (1995)



 Figure 2
Figure 2



























