« Prev Next »
The Importance of a Shared Language for Finding Information
The biggest challenge in making scientific data easy to find is the need for a common language to describe scientific terms so that a computer can scour the Internet, automatically discover relevant information, and be able to compare this information to similar data from other sources. At minimum, the languages used by different information resources need to have a common dictionary of terms that all scientists can agree on, together with a list of synonyms. In this regard, a thesaurus can go a long way toward helping software recognize similar concepts. This is the strategy that the National Cancer Institute took in 1999 when it created the NCI thesaurus (Fragoso et al., 2004) to make the knowledge contained within its cancer database more useful for computation. But such a controlled vocabulary is not enough. Without a shared language for reporting scientific findings, as well as knowledge regarding the ways in which different concepts are related, your search for "bud development" might not find a journal article describing a drug that affects gene expression during mouse limb morphogenesis, even though the result has significant applications to human disease.
An Ontology can be a Valuable Tool for Organizing Knowledge
An even more useful tool for uniting language is an ontology. Ontologies are formal ways of organizing knowledge. Traditionally studied in philosophy, ontologies have also permeated math, physics, and in the last few decades, biology. In addition to encoding a dictionary and thesaurus for a collection of words, these devices relate concepts to one another through logically defined relationships. Ontologies can be created to capture anything, although a single ontology is typically limited to a single area of knowledge, such as anatomy, cellular processes, environmental factors, and so on. For example, an ontology about experimental techniques might contain the knowledge that both "transformation" and "transfection" are "techniques that introduce exogenous DNA into cells." Whereas a thesaurus might only indicate that "transformation" and "transfection" are synonyms for a DNA introduction technique, this ontology would capture the knowledge that the two terms actually refer to specific variations of the more general technique, and that these variations are different because of the cell type they are performed on. With the knowledge encoded in the ontology, a software search engine could then scan multiple resources that tag data with these ontological terms and retrieve any protocol that describes introducing exogenous DNA into cells, regardless of the cell type involved, or whether the author used the term "transformation" or the term "transfection."
Since 1998 the Gene Ontology (GO) project (Gene Ontology Consortium, 2000) has attempted to bridge the gap between different biological communities by developing three ontologies to classify gene products: the cellular component, molecular function, and biological process ontologies. Compiled by a consortium of leading biologists, the terms in these ontologies classify gene products by where they act in the cell, what the individual products actually do, and what processes these products are part of, respectively. In addition to the GO, there are also many that classify concepts for other biological domains, such as organismal anatomy, development, experimental details, phenotype, the environment, and more. All of these recent advances are outgrowths of the Linnaean taxonomic classification system, which revolutionized biological information exchange when it was introduced.
Basic Principles of Ontologies
Logicians view an ontology as a graph of information, with terms (concepts) as nodes of the graph and relationships as the links that connect the terms. Many relationships are directed, meaning that they are only true in one direction (e.g., a nucleus is part of a cell, but a cell is not part of a nucleus); because of this, ontologies are often hierarchical in structure. The relationships used in an ontology are not predetermined, so any real-world relationship can be logically defined and used to connect terms and reflect reality. This makes ontologies a flexible framework for modeling many different kinds of data.
There are two basic relationship types used by many ontologies (Smith et al., 2005): is_a and part_of. To illustrate the different relationship types, let's consider two specific ontologies, the Zebrafish Anatomy (ZFA) and the GO.
The is_a Relationship
The part_of Relationship
Other Ontology Relationships
Inference and Transitivity
The use of defined relationships between ontological terms makes it possible to use logic to discover new information. Terms can be linked together either directly via asserted relationships or indirectly via inferred relationships. An asserted relationship is a direct relationship between two nodes (e.g., A is_a B). In contrast, inferred relationships are found by drawing a connection between two nodes across the intervening nodes and relationships, which often include multiple relationship types. Using the rules that govern the transitivity of relationships in the ontology, novel inferred relationships can then be discovered by traversing the asserted relationships.
The is_a relationship is transitive: If A is_a B, and B is_a C, then A is_a C. For example, because the heart is_a organ, and an organ is_a anatomical part, then the heart is_a anatomical part (Figure 1a).
The part_of relationship is also transitive; here, the rule states that if A is part_of B, and B is part_of C, then A is part_of C. Thus, because telophase is part_of M phase, which is part_of the cell cycle, then telophase is part_of the cell cycle (Figure 1b).
Additionally, the develops_from relationship is transitive over part_of, meaning that if A develops_from B, and B is part_of C, then A develops_from C. For instance, in Figure 1a, because the zebrafish brain develops_from the neural tube, which is part_of the ectoderm, it can be implied that the brain develops_from the ectoderm. However, part_of is not transitive over develops_from, because while the cerebellum is part_of the brain and the brain develops_from the neural tube, the cerebellum is not part_of the neural tube.
The Ontology Development Life Cycle
The development and maintenance of ontologies is truly a community effort. Experts in the field usually create an initial version of an ontology; the content and structure then grow and evolve as users discover areas that need improvement. Errors in term definitions and relationships, missing synonyms, vague terminology, and a lack (or surplus) of detail may be flagged by users. There may also be gaps in the ontology due to a lack of knowledge or because of new research. These problems are reported back to the individuals who maintain the ontology, and amendments are made to the active ontology. The incorporated changes are then released to users, and the cycle continues. GO and other biological ontologies exemplify the collaborative nature of ontology development. These ontologies are engineered not only by biologists and/or medical doctors themselves, but also by special workshops in which community experts are brought in to clarify and expand the branches of an ontology, or to correct cases in which an ontology has "diverged from current usage and understanding in the field" (Diehl et al., 2007).
Gaps in the scientific domains modeled by current ontologies trigger new ontologies to be developed. Because much scientific knowledge is already encoded in existing ontologies, the development of new ontologies can often be expedited by creating links between existing ontological terms. For example, a disease ontology that defines a particular type of breast cancer as being caused by a mutation in BRCA1 could link to the Gene Ontology, which has a term for a BRCA1-protein complex. Note that duplication of concepts in different ontologies should be avoided, where possible, so that the knowledge contained in each ontology is orthogonal to the rest. This practice helps prevent confusion for users of the ontologies, and it also avoids redundancy in the knowledge models.
Biocuration and Using Ontologies for Discovery
Biocuration is a growing field in which expert curators read and distill scientific findings from the published literature by utilizing ontological terms to "tag" them (Howe et al., 2008). One important and challenging curatorial task is relating a gene (and/or its gene products) to the anatomical location where it is expressed, its molecular function, and/or the biological process it participates in (Hill et al., 2008). Here, curators must identify important results from genetic, molecular, and biochemical laboratory experiments that are reported in the literature and then annotate the gene with the appropriate ontological terms.
Nonautomated annotation efforts using the Gene Ontology, as well as other anatomically specialized ontologies, are currently being performed at several model organism databases, such as VectorBase (disease-carrying insects), ZFIN (zebrafish), and SGD (yeast). The combination of these annotations, together with the knowledge contained in the ontology, allows striking results to be achieved. For example, one of the goals for research on model organisms is to "model" the biology of humans; thus, merging the phenotypic data from different model species and performing statistical analysis has the power to provide insights into human health and disease.
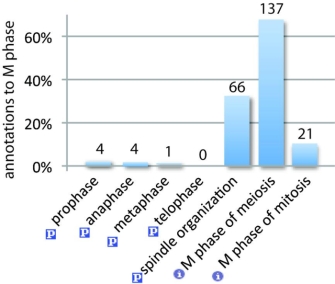
The AmiGO browser is one example of a tool that merges data from many different model organism databases, such as those for yeast, flies, and mice. With this tool, a user can browse and compare the annotations of genes from different organisms because the annotations are made with a common ontology (GO). The relationships and structure of the ontology make a question such as "What are all of the C. elegans genes that are involved in the cell cycle?" easy to answer, because a computer can deduce that, due to the ontological relationships and their transitivity rules, any annotations made to the parts of the cell cycle are also annotations to the cell cycle itself. For instance, browsing AmiGO shows that four annotations were made directly to "anaphase," with the specific gene names shown in Figure 1b. Only three annotations were made to the term "cell cycle" itself; the rest of the annotations propagate "up" to cell cycle from its child terms, and together with the inferred annotations, they result in a total of 286 annotations. Figures 1b and 2 show the distribution of genes annotated to the children of "M phase."
Some automated curation methods, such as those using natural language processing (NLP), can make a first pass at tagging the literature. For example, the Neurocommons Pilot Project has computationally annotated more than 300,000 neuroscience publications by applying ontological tags to each abstract, then publishing the annotations in a standard data format. The tags include gene names, along with functions or processes that those genes might participate in (e.g., activation, interaction, gene expression). These automated techniques, together with expert curation, provide breadth and depth to the biological annotation. They also bring previously disconnected data together into a rich, searchable format. Specialized "intelligent" search engines are also being developed that utilize ontologies to retrieve and organize the most relevant results, which will reduce the time spent manually sifting through them.
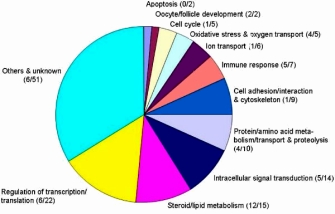
Yet another powerful analysis technique is to compare the annotations for a set of genes that are known to be expressed under one condition with those expressed under another condition(s), by asking questions such as, "What are the differences in the biological processes that these genes are involved in?" This type of analysis is a common way to analyze comparative expression microarrays. An example of the results of this sort of analysis is shown in Figure 3. Here, Vanselow et al. (2008) isolated differentially expressed transcripts from two different mutant mouse lines known to vary in their fertility, and they mapped the involved genes to numerous biological processes in GO. Many were female reproductive processes, including folliculogenesis, ovulation, and luteinization, from which the researchers concluded that the genes might play a role in increasing the ovulation number in mutant mice, thus making these animals more fertile.
To return to your original quest—finding the genes involved in bud development—you can now see how ontologies, and the annotations made with them, can greatly facilitate research. By ensuring that everyone is speaking the same language, ontologies help break down the communication barriers that hinder scientific discovery. The relationships that connect biological knowledge provide an extremely powerful and versatile system that makes sharing data easier and more rewarding, and that encourages different scientific disciplines to collaborate. In an era in which online activity is becoming increasingly social and collaborative, it is only natural that science should follow suit. With a little bit of effort to encode scientific findings in a structured, knowledge-enhanced way using ontologies, researchers can make data available for discovering novel connections, which maximizes the overall scientific impact of all research efforts.
References and Recommended Reading
Bard, J., & Rhee, S. Ontologies in biology: Design, applications, and future challenges. Nature Reviews Genetics 5, 213–222 (2004) (link to article)
Bodenreider, O., & Stevens, R. Bio-ontologies: Current trends and future directions. Briefings in Bioinformatics 7, 256–274 (2006)
Carbon, S., et al. AmiGO: online access to ontology and annotation data. Bioinformatics. (2008)
Diehl, A. D., et al. Ontology development for biological systems: Immunology. Bioinformatics 23, 913–915 (2007)
Fragoso, G., et al. Overview and utilization of the NCI Thesaurus. Comparative and Functional Genomics 5, 648–654 (2004)
Gene Ontology Consortium. Gene ontology: Tool for the unification of biology. Nature Genetics 25, 25–29 (2000) (link to article)
Hill, D. P., et al. Gene ontology annotations: What they mean and where they come from. BMC Bioinformatics 9, S2 (2008)
Howe, D., et al. Big data: The future of biocuration. Nature 455, 47–50 (2008) (link to article)
Noy, N. F., & McGuinness, D. L. Ontology Development 101: A Guide to Creating Your First Ontology. http://protege.stanford.edu/publications/ontology_development/ontology101-noy-mcguinness.html
Smith, B., et al. Relations in biomedical ontologies. Genome Biology 6, R46 (2005)
Smith, B., et al. The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration. Nature Biotechnology 25, 1251–1255 (2008) (link to article)
Vanselow, J., et al. Expression profiling of a high-fertility mouse line by microarray analysis and qPCR. BMC Genomics 9, 307 (2008)



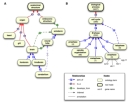
 Figure 1
Figure 1