« Prev Next »
Computer databases are an increasingly necessary tool for organizing the vast amounts of biological data currently available and for making it easier for researchers to locate relevant information. In 1979, the Los Alamos Sequence Database was established as a repository for biological sequences. In 1982, this database was renamed GenBank and, later the same year, moved to the newly instituted National Center for Biotechnology Information (NCBI), where it lives today. By the end of 1983, more than 2,000 sequences were stored in GenBank, with a total of just under 1 million base pairs (Cooper & Patterson, 2008).
At about the same time, a joint effort between NCBI, the European Molecular Biology Laboratory (EMBL), and the DNA Databank of Japan (DDBJ) created the International Nucleotide Sequence Database Collaboration (INSDC) to collect and disseminate the burgeoning amount of nucleotide and amino acid sequence data that was becoming available. Since then, the INSDC databases have grown to contain over 95 billion base pairs, reflecting an exponential growth rate in which the amount of stored data has doubled every 18 months (Figure 1). The advent of next-generation sequencing technologies, metagenomics, genome-wide association studies (GWAS), and endeavors such as the 1000 Genomes Project will only increase the tremendous volume and complexity of this and other sequence data collections (Siva, 2008).
The sheer volume of the raw sequence data in these repositories has led to attempts to reorganize this information into various kinds of smaller, specialized databases. Such databases include various genome browsers, model organism databases, molecule- or process-specific databases, and others. To get an understanding of the growth of these resources, one need only look at the annual database issue of the journal Nucleic Acids Research. In one of the first database issues—the one in which GenBank is described—only a few dozen databases are listed (Bilofsky et al., 1986). In contrast, the latest database issue describes over 1,000 genomics databases and tools (Galperin, 2008). However, even this list of resources is only part of the overall picture. Today, it appears that there are upwards of 3,000 distinct genomic resources, tools, and databases publicly available on the Internet. This article outlines the current state of such databases, reviewing some of the databases available for use, various issues concerning the growth of these databases, and the number and sheer size of these resources.
Sequence Data Repositories
As previously mentioned, the INSDC is a collaboration of NCBI's GenBank in the U.S., EMBL in Europe, and the DDBJ in Japan. Each of these databases accepts direct submissions of biological sequences from individual researchers, from sequencing projects, and from patent applications from around the world. Sequences are entered into the database and given a unique identification or accession number. These submitted entries are stored in a "library" of records, and each entry is "owned" by—and can only be updated by—its submitter. The data integrated in these entries include the submitter's name, the originating organism, the definition, the actual sequence, related references, and more. (Examples of IDs and and entries are accessible through these links.) The submitted entries are then shared across the three repositories on a daily basis, and releases of the data are made regularly. This has been a boon to the research community, facilitating the sharing of sequence data and allowing the advancement of research. These sequence repositories have become the universal, comprehensive, and authoritative resources for the exponentially growing amount of sequence data currently available to researchers.
However, there are challenges associated with these repositories. For one, exponential growth makes it difficult to maintain accuracy and accessibility across the three databases. Although researchers are able to update sequences they have submitted to GenBank and other repositories, a large portion of the stored data may be incorrect or incomplete due to the volume of the submitted information and the nature of research (e.g., researchers move on to other projects, mistakes in the original data go unnoticed, etc.). There are also issues of duplication with minor variations and redundancy. Because of this, several programs and efforts have been developed to help correct and curate sequence data. (Pruitt et al., 2005). RefSeq is a database developed and maintained at NCBI that aims to provide a scientist-curated nonredundant set of biological sequences. Here, the data are propagated from sequences deposited within the GenBank database and then carefully annotated using community collaboration, automated computer annotation, and NCBI staff curation. This provides a reference set of well-curated sequences. Related efforts include (a repository for high quality, frequently updated, manual annotation of vertebrate finished genome sequences), , "a collaborative effort to identify a core set of human and mouse protein coding regions that are consistently annotated" [National Center for Biotechnology Information, 2008]), and other projects that utilize the INSDC archival database sequences and build on them to create curated, accurate sequence data. Note, however, that these projects cannot tackle every species and sequence in each repository, especially because sequence data is produced faster than researchers can annotate and organize it.
Another challenge associated with genome repositories is that repository data rarely have context or annotation, nor are they organized in a manner that facilitates research. For example, a gene record might be linked to references for what the gene does, but images showing localization in the cell or primary data showing potential protein modifications might only be accessible in research papers and not databases. This means that much of the data are difficult to access and utilize. Given this situation, database developers have taken the data from GenBank and repurposed it to their specific aesthetic, project history, annotation, and community needs. As a result, there are currently many databases and strategies for presenting and providing access to genomic data. The main categories of such databases are described in the sections that follow.
General Genome Browsers
One highly useful answer to the needs of many researchers is the development of general genome browsers. Three successful examples include the University of California, Santa Cruz (UCSC) Genome Browser, EBI's Ensembl, and NCBI's MapViewer (Karolchik et al., 2008; Flicek et al., 2008; Wheeler et al., 2008b). These browsers repackage genome and gene annotation data sets from GenBank and other subject-specific databases to provide a genomic context for individual genome features, such as genes or disease loci. Furthermore, not only do these browsers provide genomic context, but they also allow users to see common formats between diverse species so that information is more easily viewable and extractable. For example, a user can search for a specific region of a genome, such as a disease gene, and the sequence and attendant annotation will be displayed visually. This display will then link out to additional data and databases for further study, as well as back to repository sources for the original data (Figure 2).
This framework, based on the official genomic sequence, also permits the addition of new data and data types organized according to the sequence. For instance, new ENCODE data that maps regulatory elements and many other DNA elements can be easily placed on this existing structure to enhance researchers' understanding of any given genomic region (ENCODE Project Consortium, 2007). These genome browsers also offer advanced searching capabilities, such as Table Browser (UCSC) and BioMart (Ensembl), which allow researchers to obtain and analyze data in a highly customized fashion (Karolchik et al., 2005; Fernandez-Suarez & Birney, 2008). These customized searches and data downloads can be used as input for further bioinformatics analyses or experiments to verify computational predictions in the lab.
Species- and Taxa-Specific Databases
The aforementioned general genome browsers include genomes and annotations for dozens of species in their databases, and they give researchers an excellent first stop to analyze a breadth and depth of data that otherwise might be difficult to obtain. Nonetheless, many researchers require even deeper information about various species' genomes.
Thus, hundreds of species-specific or taxa-specific genome databases have been developed by various research communities and groups. These databases and browsers are, more often than not, publicly available for researchers to access and use. The databases employ a variety of interfaces and tools to serve their communities of researchers. They can be quite extensive, and they may also be deeply curated. Such databases are often accurate, up-to-date sources for annotations because they employ manual curation of relevant literature and integrate it into gene annotation data. They also incorporate more species-specific data types than some of the generalized genome databases. Some examples of these species- and taxa-specific resources are listed in Table 1.
Table 1. Genomics Databases
Standardized Genome Database Tools: GMOD
Because of historical, biological, and practical reasons, data are not completely consistent between species genomes and research projects. Model organism databases often have unique schemas that are not easily comparable to those used in databases for other species. Indeed, the terminology, analysis techniques, and importance attached to different sequence elements and annotations can be quite different across databases. The genome browsers mentioned above are one solution to this problem. However, another option has emerged to provide deeper and broader data for individual species' genomes, as well as increased standardization that allows for better cross-species comparisons and greater ease of use.
In particular, the consortium has worked on the development of an open-source standard database and set of visualization tools to make querying, browsing, and using genome databases similar for all species. GMOD has collaboratively developed a set of tools and database schema that include an annotation editor (Apollo), a genome browser (GBrowse), pathway tools, an advanced search capability (BioMart), a biological database schema (Chado), and additional resources that allow species research communities to develop databases that are standard and compatible across genomes. The goal of these efforts is to facilitate research and comparative studies.
Many species- and taxa-specific genome databases have made use of this standard, open-source set of database tools. The RGD, TAIR, Gramene, FlyBase, MGI, SGD, and WormBase databases are just a few. Sometimes, these tools are the main foundation of a database; in other cases, they supplement existing databases. Organisms with smaller research communities can also use these handy tools to create the annotation, visualization, and query options they need. As the rate of genome sequencing continues to increase thanks to new technologies, the GMOD tools may prove to be a boon for researchers who need to better explore their sequences of interest. Note that these tools can support many data types. In fact, there are many diverse and creative examples of ways in which the GMOD tools can be used, such as the (HGSDD), human variation data, and even personal genomics in the form of Watson's genome (Cheung & Estivill, 2003; International HapMap Consortium, 2003; Wheeler et al., 2008a). You can find a large and varied list of resources that use these tools by accessing the GMOD website.
Subject-Specific Databases
In addition to species- or genome-oriented databases, there are also databases organized by almost any biological data category one can imagine. For example, there are databases specifically for protein domain information (Pfam) and protein structure information (PDB). There are also repositories and databases of expression data, such as NCBI's Gene Expression Omnibus (GEO) and EBI's ArrayExpress (Berman & Westbrook, 2000; Barrett et al., 2006; Parkinson et al., 2007). In addition, GWAS databases such as dbGaP and HuGE Navigator are emerging (Yu et al., 2008). The list of subject-specific databases is quite large—as mentioned earlier, there are over 3,000 such resources—and the variety of these "focused" databases is as unlimited in scope as the data they contain. Nonetheless, this large number of species- and subject-specific databases, though extremely useful, can lead to its own issues of redundancy and lack of integration.
Solutions to the Current Challenges of Accuracy and Curation
All of the aforementioned resources, from the respositories to the genome databases and subject-specific databases, are increasingly faced with the challenge of ensuring accurate data and efficiently managing and curating that data. Recently, several solutions have been proposed (Waldrop, 2008; Howe et al., 2008). These solutions range from a greater focus on the education of database biocurators in learning institutions and the standardized inclusion of sequence data and references in publications to "community curation." One community curation solution envisions a sort of "wikification" of data update and curation, in which research communities curate their databases themselves. This has been proposed for repositories, specifically GenBank, as well as for focused resources, such as model organism databases (Pennisi, 2008; Salzberg, 2007).
GenBank has resisted this "wikification" proposal for various reasons, feeling that the current system allows an authoritative repository and a database of record and that community editing might diminish this strength. Additionally, there are already programs and efforts aimed at correcting and curating the sequence data, such as RefSeq (mentioned earlier in this article). As for model organism databases, there are currently several relatively successful efforts at community curation and annotation, including the Daphnia Genomics Consortium wiki and several other extensive undertakings. However, these efforts are hampered by factors such as the reliability of curation, the lack of incentives for researchers to contribute, and more. As the authors of a recent paper in Nature suggest, "To date, not much of the research community is rolling up its sleeves to annotate" (Howe et al., 2008).
Discussion and Future Challenges
Various efforts at building archives, databases, and analysis tools have proven successful at facilitating a better understanding of the genomes of multiple species. They have offered researchers authoritative repositories, contextual information, and curated data as a method of handling the exponentially growing amount of sequence data. Although these resources have been useful and have solved many issues, they will continue to face new types and ever-growing amounts of data that will exacerbate the challenges with which the research community is already faced. For example, genome-wide association studies will generate an enormous amount of data that will provide insight into the multifactorial genetic origins of disease, evolution, and more.
These data have also created new challenges related to the development of methods for visualizing and searching information. Recently, unforeseen privacy issues have required that large datasets be removed from public databases (Couzin, 2008; Zerhouni & Nabel, 2008) because personal genetic information could be associated to individuals. Metagenome sequencing projects, which analyze communities of genomes instead of individual genomes, are also creating large, complicated data sets that require unique tools and databases (Markowitz et al., 2008a; National Research Council Committee on Metagenomics, 2007).
Lastly, but importantly, the growing number of genome databases, analysis tools, and other resources available on the web has made it daunting for researchers to use these resources effectively. Even with efforts toward standardization and documentation, researchers continue to find it difficult to locate and learn to use these resources (Collins & Green, 2003). Solutions involving advanced, life-long training on the use and access of specific resources must be found. Next-generation sequencing and personal genomics will further burden efforts in this arena.
In spite of the challenges that have arisen with the growth of data and databases, the rewards and opportunities provided by this information have proven fruitful. Today, there is a wealth of data that was undreamed of just a couple of decades ago, enabling new discoveries and uncovering new relationships between different disciplines. The authors often joke to their students that if these resources had been available when they were in graduate school just 15 years ago, it would have taken them months—not years—to complete their degrees. With the growth of available data and resources in the next few years, amazing discoveries will continue to be made if the scientific community can meet the challenge.
References and Recommended Reading
Barrett, T., et al. NCBI GEO: Mining tens of millions of expression profiles—Database and tools update. Nucleic Acids Research 35, D760–D765 (2006)
Berman, J., & Westbrook, Z. The Protein Data Bank. Nucleic Acids Research 28, 235–242 (2000)
Bilofsky, H. S., et al. The GenBank genetic sequence databank. Nucleic Acids Research 14, b1–b4 (1986)
Bult, C., et al. The Mouse Genome Database (MGD): Mouse biology and model systems. Nucleic Acids Research 36, D724–D728 (2008)
Cheung, J., & Estivill, X. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biology 4, R25 (2003)
Collins, F., & Green, E. A Vision for the future of genomics research. Nature Genetics 422, 835–847 (2003) (link to article)
Cooper, E., & Patterson, I. The legacy of GenBank: The DNA sequence database that set a precedent. 1663: The Los Alamos Science and Technology Magazine, http://www.lanl.gov/news/index.php/fuseaction/1663.article/d/200808/id/14273 (2008)
Couzin, J. Genetic privacy: Whole-genome data not anonymous, challenging assumptions. Science 321, 1278 (2008)
ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007) doi: 10.1038/nature05874 (link to article)
Fernandez-Suarez, X., & Birney, E. Advanced genomic data mining. PLoS Computational Biology 4, e1000121 (2008)
Finn, R., et al. The Pfam protein families database. Nucleic Acids Research 36, D281–288 (2008)
Flicek, P., et al. Ensembl 2008. Nucleic Acids Research 36, D707–D714 (2008)
Galperin, M. Y. The molecular biology database collection: 2008 update. Nucleic Acids Research 36, D2–D4 (2008)
Hong, E. L., et al. Gene ontology annotations at SGD: New data sources and annotation methods. Nucleic Acids Research 36, D577–D581 (2008)
Howe, D., et al. Big data: The future of biocuration. Nature 455, 47–50 (2008) (link to article)
International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003) (link to article)
Karolchik, D., et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Research 33, D454–D458 (2005)
———. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Research 36, D773–D779 (2008)
Lawson, D., et al. VectorBase: A home for invertebrate vectors of human pathogens. Nucleic Acids Research 35, D503–D505 (2007)
Liang, C., et al. Gramene: A growing plant comparative genomics resource. Nucleic Acids Research 36, D947–D953 (2008)
Markowitz, V., et al. IMG/M: A data management and analysis system for metagenomes. Nucleic Acids Research 36, D534–D538 (2008a)
———. The Integrated Microbial Genomes (IMG) system in 2007: Data content and analysis tool extensions. Nucleic Acids Research 36, D528–D533 (2008b)
Maxam, A. M., & Gilbert, W. A new method for sequencing DNA. Proceedings of the National Academy of Sciences 74, 560–564 (1977)
National Center for Biotechnology Information. CCDS database, http://www.ncbi.nlm.nih.gov/CCDS (2008)
National Research Council on Metagenomics. The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet (Washington, DC, National Academies Press, 2007)
Parkinson, H., et al. ArrayExpress: A public database of microarray experiments and gene expression profiles. Nucleic Acids Research 35, D747–D750 (2007)
Pennisi, E. Proposal to "wikify" GenBank meets stiff resistance. Science 319, 1598–1599 (2008)
Pruitt, K. D., et al. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence of database genomes, transcripts, and proteins. Nucleic Acids Research 33, D501–D504 (2005)
Rogers, A., et al. WormBase 2007. Nucleic Acids Research 35, D612–D617 (2007)
Salzberg, S. Genome re-annotation: A wiki solution? Genome Biology 8, 102 (2007)
Sanger, F., & Coulson, A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. Journal of Molecular Biology 94, 441–448 (1975)
Siva, N. 1000 Genomes Project. Nature Biotechnology 26, 256 (2008) (link to article)
Sprague, J., et al. The Zebrafish Information Network: The zebrafish model organism database. Nucleic Acids Research 34, D581–D585 (2005)
Swarbreck, D., et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Research 36, D1009–D1014 (2008)
Twigger, S., et al. Rat Genome Database update 2007: Easing the path from disease to data and back again. Nucleic Acids Research 35, D658–D662 (2007)
Waldrop, M. Big data: Wikiomics. Nature 455, 22–25 (2008)
Watson, J. D., & Crick, F. H. A structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953) (link to article)
Wheeler, D., et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 17, 872–876 (2008a) (link to article)
Wheeler, D., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 36, D13–D21 (2008b)
Wilming, L., et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Research 36, D753–D760 (2008)
Wilson, R., et al. FlyBase: Integration and improvements to query tools. Nucleic Acids Research 36, D588–D593 (2008)
Yu, W., et al. A navigator for human genome epidemiology. Nature Genetics 40, 124–125 (2008) (link to article)
Zerhouni, E., & Nabel, E. Protecting aggregate genomic data. Science 321, 1 (2008)



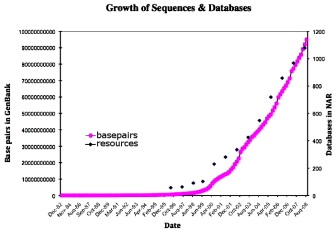
 Figure 1: The phenomenal growth of sequence data in GenBank is challenging to manage, and continues unabated.
Figure 1: The phenomenal growth of sequence data in GenBank is challenging to manage, and continues unabated.



























