Abstract
Development of apple (Malus domestica) cultivars resistant to fire blight, a devastating bacterial disease caused by Erwinia amylovora, is a priority for apple breeding programs. Towards this goal, the inactivation of members of the HIPM and DIPM gene families with a role in fire blight susceptibility (S genes) can help achieve sustainable tolerance. We have investigated the genomic diversity of HIPM and DIPM genes in Malus germplasm collections and used a candidate gene-based association mapping approach to identify SNPs (single nucleotide polymorphisms) with significant associations to fire blight susceptibility. A total of 87 unique SNP variants were identified in HIPM and DIPM genes across 93 Malus accessions. Thirty SNPs showed significant associations (p < 0.05) with fire blight susceptibility traits, while two of these SNPs showed highly significant (p < 0.001) associations across two different years. This research has provided knowledge about genetic diversity in fire blight S genes in diverse apple accessions and identified candidate HIPM and DIPM alleles that could be used to develop apple cultivars with decreased fire blight susceptibility via marker-assisted breeding or biotechnological approaches.
Similar content being viewed by others
Introduction
Breeding perennial trees and woody crops is a challenge due to their long juvenile periods, slow growth rates, and high heterozygosity. A combination of next-generation sequencing and marker-assisted selection (MAS) can overcome many of these challenges and reduce the time needed for each selection cycle1,2,3,4,5,6. Given the economic impact, fruit quality and, more recently, disease resistance have been the major focus for apple breeders. The introgression of disease resistance into commercially preferred backgrounds is often problematic, given that the majority of known major effect resistance loci exist within wild species7. Wild Malus species often have poor fruit quality and it can take up to 25–30 years to fully recover fruit quality and eliminate linkage drag of undesirable alleles in a Malus domestica background8. Self-incompatibility and high heterozygosity also make breeding outside of M. domestica not feasible in many cases9. Thus, within this context, developing sequence-based markers and using them in targeted MAS can greatly reduce the length of breeding schemes.
The molecular markers linked with traits can be identified using bi-parental QTL (Quantitative Trait Loci) mapping, genome-wide (GWAS) or candidate gene-based association mapping approaches. The GWAS approach is based on the use of several thousand markers evenly spaced across the whole genome that can be implemented to identify QTLs. Candidate gene-based association mapping uses molecular markers from genes with known trait-associated functions from literature to identify the genetic variants that most affect the phenotype. In Malus, this approach has been used to define impacts on the depolymerization of pectin and changes in the resulting fruit texture of variants in the Md-PG1 gene identified by resequencing10. These gene-based association studies highlight the importance of identifying specific markers for trait-specific MAS.
Fire blight, caused by the gram-negative bacterium Erwinia amylovora, has a devastating economic impact on apple, pear, and quince production worldwide6,7,11,12. The limited resistance to fire blight in modern commercial apple cultivars has led to increased reliance on antibiotics, growth inhibitors, and crop insurance claims13. In different Malus species, several major and minor effect fire blight resistance QTLs have been identified. For example, major resistance QTLs were found in M. robusta, M. floribunda, M. ‘Evereste’, and M. fusca on linkage groups 3, 12, 12, and 10, respectively14,15,16. The challenges of conventional apple breeding make these QTLs most useful for marker-assisted breeding or genome editing. Studies have also identified minor to moderate-effect fire blight resistance loci within M. domestica. For example, QTLs have been identified on ‘Fiesta’, ‘Prima’, and ‘Discovery’ on linkage group 7, and in ‘coop16’ and ‘coop17’ on linkage groups 2, 6, and 15, respectively17,18,19. Due to the rapid rate of evolution of E. amylovora, these resistance QTLs, particularly major effect QTLs, can be overcome rather quickly20,21. For example, a non-synonymous SNP in the AvrRpt2EA effector gene could overcome major fire blight resistance gene FB_MR5, derived from an accession of Malus robusta20,21.
Susceptibility (S) genes can provide a broad-spectrum and more durable alternative to narrow-spectrum and pathogen-specific resistance genes to develop cultivars with reduced disease susceptibility. Resistance genes typically recognize specific pathogen-encoded avirulence (Avr) proteins to induce the resistance reponse. Whereas, S genes facilitate host recognition and compatibility by encoding negative regulators of immune signaling, as well as providing for successful pathogen penetration, establishment and proliferation, therefore, reducing their activity can lead to a more resistant plant22. The use of susceptibility genes in breeding biotic stressors has been relatively limited. X5, Xa13, and eIF4G in rice as well as eIF4E in barley, pepper, melon, pea, and lettuce are a few S genes that have been targets of breeding efforts23,24,25,26,27,28,29,30,31,32. Susceptibility genes are well suited for genome editing, as targeted knockouts via CRISPR/Cas9 or TALENs can subtract the genetic contribution of S genes, decreasing the disease susceptibility of the plant. A knockout of the grapefruit S gene CsLOB via CRISPR/Cas9 resulted in reduced infection by the citrus canker pathogen (Xanthomonas citri ssp.) compared to the wild type (Duncan grapefruit; Citrus × paradisi)33. For Malus, two susceptibility genes, HIPM and DIPMs (DIPM1-4) have been identified as key regulators of establishment and proliferation of E. amylovora34,35. HIPM (HrpN-interacting protein from Malus) interacts with the primary class of effector proteins (HrpN) in E. amylovora that initiate infection, the production of reactive oxygen species, and the induction of hypersensitive response in the host36,37,38. HIPM is a transmembrane protein of 60 amino acids and was demonstrated to be expressed constitutively in apple tissues, particularly in flowers, the most critical tissue for E. amylovora infection36. The reduction in expression via RNAi of HIPM in the cultivar ‘Galaxy’ resulted in decreased fire blight susceptibility in the transgenic plants35,38. Similarly, DIPMs (DspA/E-interacting proteins from Malus) are apple proteins encoded by a family of four genes (DIPM1-4, with length of 666, 676, 665 and 682 amino acids, respectively) and are a close match to transmembrane leucine-rich repeat receptor-like kinase sequences in other organisms39. DIPMs are expressed in young shoots and their transcript level increases upon E. amylovora infection, except for DIPM139. Moreover, using yeast two-hybrid and pull-down assays, Meng and colleagues39 demonstrated that the kinase domains of DIPM1, 2, and 4 interact physically and specifically with the E. amylovora effector protein DspA/E. On the contrary, DIPM3 is not subjected to this interaction. Within this framework, the same authors preliminarily concluded that DIPMs may act as susceptibility factors during the apple-E. amylovora interaction. Recently, it was shown that the inactivation of MdDIPMs via RNAi or CRISPR/Cas9 in the commercial cultivars ‘Gala’ and ‘Golden Delicious’ altered the disease phenotypes of the edited plants and reduced their susceptibility levels to E. amylovora strain Ea27334,40, thus effectively confirming the involvement of DIPMs in apple susceptibility.
This study identifies putative non-functional alleles of HIPM and DIPM genes from publicly available apple genomes, investigates the genomic diversity of HIPM and DIPM genes in Malus germplasm collections, evaluates a diverse set of Malus accessions for fire blight resistance in the greenhouse, and uses a candidate gene-based association mapping approach to identify SNPs (Single nucleotide polymorphisms) in these genes showing significant association with fire blight susceptibility phenotypes.
Materials and methods
Plant material
A total of 238 apple accessions, from four different sources, were included in this study (Supplementary Table S1). Collection 1 had a total of 145 Malus accessions with whole-genome resequencing data. These are from three different sources, the majority from M. domestica, M. sieversii, and M. sylvestris that were previously sequenced41, 67 accessions that were sequenced separately42, and eight further accessions of M. sieversii and M. sylvestris sequenced in this study (unpublished data). Collection 2 had 93 accessions from 26 Malus species that were selected from the US National Malus Collection (Geneva, NY, USA) based on fire blight resistance ratings available at Germplasm Resources Information Network (GRIN) (www.ars-grin.gov), 70 of which overlap with collection 1. Information regarding the accessions, including Plant Introduction (PI) number, name, species, origin, ploidy level and fire blight resistance ratings (available at GRIN) are reported in Supplementary Table S1.
The apple accessions from collection 2 were evaluated for fire blight resistance and susceptibility in a controlled greenhouse experiment. Three replicates of each accession were grafted via whip and tongue by interlocking corresponding cuts in the scion wood and ¼ inch ‘M.7’ rootstocks and potted in D40 large Deepots (Stuewe and Sons, Tangent, OR) in Sungrow #8/Fafard #2 (Sungrow Horticulture Agawam, MA) soil mix. The plants were grown in a greenhouse at a temperature between 21 and 24 °C with 12 h supplemental light and were watered and fertilized as needed.
Genomic sequence and primers for HIPM and DIPM genes
The HIPM and DIPM gene sequences were detected in the ‘Golden Delicious’ double-haploid apple genome (GDDH13 v1.1)43 using a blast search with previously reported sequences36,39. The detected gene sequences were used as templates to design ~ 150 base pair amplicons to cover entire gene sequences using a custom python script (available at https://bitbucket.org/michelettd/ampseq-primer-design). The primers were designed using Primer3 (https://primer3.ut.ee, version 4.1.0) to amplify 150 ± 10 bp of fragments of the coding sequences from HIPMs and DIPMs. Besides the use of default parameters, all the primers were blast-searched against the GDDH13 v1.143 reference genome to exclude putative repeat regions. The known SNPs were masked in the target regions to avoid annealing problems. On average, five primer pairs were selected for each gene in order to amplify gene fragments at intervals of approximately 300–600 base pairs via AmpSeq genotyping (see below) (Supplementary Table S2).
Genomic DNA extraction and AmpSeq genotyping
Total genomic DNA from Malus accessions of Collection 2 (Supplementary Table S1) was extracted from young fresh leaves using the Wizard Genomic DNA Purification Kit (Promega, USA), according to the manufacturer's instructions. The concentration of extracted DNA was analyzed with a NanoDrop Spectrophotometer (Thermo Fisher Scientific, Gand Island, NY, USA) and the quality of DNA samples was assessed by running a 0.8% agarose gel at 80 V for 1.5 h. Genomic DNA was amplified with the previously designed and selected primer pairs (Supplementary Table S2) to amplify amplicon pools, which were sequenced on the Illumina MiSeq Platform (Illumina, San Diego, CA, USA), according to the AmpSeq protocol at the Genomics Platform Facility of Cornell University (Ithaca, NY, USA)44.
Sequence analysis and variant detection
The sequences from AmpSeq run were demultiplexed using the barcode sequences. The raw sequences from 93 AmpSeq (Collection 2) were quality assessed using fastqc tool (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and reads were trimmed and filtered to remove sequencing barcodes and low-quality bases using Trimmomatic software with the parameters: Trailing 20, Leading 20, SlidingWindow 4:15, AvgQual 20, MinLen 2545. For the remaining 145 accessions, whole-genome sequencing data were available, and the reads were filtered for quality as described above. The remaining high-quality reads were aligned against the GDDH13 v1.143 genome with burrows-wheeler aligner (BWA) with default parameters. PCR duplicate reads were removed with Picard tools43.
Variants were identified using the HaplotypeCaller function of the Genome Analysis Toolkit (GATK version 4.1) using default parameters to detect SNPs and short Indels across the HIPM and DIPM gene sequences. The final SNP dataset was partitioned into three different datasets for (1) 145 Malus samples (Collections 1), (2) 93 AmpSeq samples (Collection 2), and (3) sum of the two collections. The variants were further filtered using a mean read depth threshold > 3. The variants were annotated using the coding and protein sequences of HIPM and DIPM genes from the GDDH13 v1.143 genome. In addition, the gene annotations were obtained from the GDDH13 v1.143 genome to annotate variants using the ANNOVAR program46.
Assessment of genomic diversity
The genomic diversity across HIPM and DIPM gene sequences was determined by calculating nucleotide diversity (π) and fixation index (Fst) statistics across the three SNP datasets with the program VCFtools47. In addition, the population genetic structure was determined by performing principal component analysis (PCA) of the genetic variants with Tassel v5 software48.
Fire blight inoculation and data collection
The fire blight inoculation of grafted plants was performed with highly virulent strains of E. amylovora (E2002a, Ea273, and E4001a)49 to evaluate varying levels of resistance and susceptibility. In 2018, the population was screened with a 1:1:1 ratio of E2002a, Ea273, and E4001a at a concentration of 1 × 109 CFU/ml. In 2019, the only Ea273 strain was used at the same concentration as the prior year. The preparation of the inoculum consisted of plating the E. amylovora strain on King’s Medium B Base culture media, incubated at 27 °C for 2 days. Visible bacterial colonies were then scraped and added to 1× Phosphate Buffered Saline (PBS, pH 7.4) solution to halt growth of the bacteria. The bacterial suspension was quantified with a Smart Spec Plus Spectrophotometer (BioRad Hercules, CA) based on the optical density at 600 nm and diluted accordingly with 1× PBS to a concentration of 1 × 109 CFU/ml. The inoculation was done by cutting the youngest leaf on the newest grown shoot with sterilized scissors dipped in the inoculum. During the early infection period, the temperature and humidity of the greenhouse was raised to 25–27 °C and 75–80%, respectively. After 12 days in 2018 and 8 days in 2019, the data of the fire blight infection was collected by measuring the leaf length (LL; cm) from the scissor cut to the end of leaf blade, leaf necrosis length (LN; cm) from the scissor cut to the visible lesion on the leaf, shoot length (SL; cm) of the current year’s growth from the node of the inoculated leaf and shoot necrosis length (SN; cm), the visible necrosis in the current year’s growth from the node of the inoculated leaf.
Statistical analysis of fire blight data
Analysis of fire blight data was performed in R Version 3.6.250. The datasets from each year were analyzed to remove any outliers. In 2019, plants with only old lignified leaves available for infection were noted and filtered out from further analysis. The remaining data were filtered for the genotypes with data for at least three replications. The leaf length (LL), leaf necrosis length (LN), shoot length (SL), and shoot necrosis length (SN), evaluated as above, were used to calculate percent leaf lesion length (PLLL), percent shoot lesion length (PSLL), and percent total distance (PTD) as follows;
PSLL is described as percent shoot necrosis (PSN) or percent lesion length (PLL) in previous literature18,51. For each genotype, the mean and standard deviation of all the replications were calculated for 2018 and 2019, separately. For the traits PLLL, PSLL, and PTD, a two-way analysis of variance (ANOVA) was performed to test for significant differences in phenotype across genotypes. To assess the homogeneity of the variances across the 2 years of data and disease traits within accessions, a non-parametric Fligner–Killeen test was conducted on the PLLL, PSLL, and PTD traits. In addition to the genotypic effects, the ANOVA model also accounted for the length of the inoculated leaf as covariates as well as the interaction effect between the individual genotype and the leaf length. The disease phenotype for each year was clustered to coerce accessions into classes of resistances using the partition k-means clustering approach. To perform clustering, the infection data for the traits PLLL and PTD of each genotype was scaled to a distribution mean of zero and a standard deviation of one via a Z-score normalization. Euclidean cluster distances were calculated using the ‘k-means’ package in R Version 3.6.2 based on the algorithm of Hartigan and Wong (1979). The number of clusters was determined using the ‘NbClust’ package in R Version 3.6.2 based on 30 unique indices that determine the optimal number of clusters representing a dataset50,52.
Association between fire blight susceptibility and genomic variations
The fire blight phenotype data and the corresponding SNP genotypic data was used to conduct iterative marker-trait associations. The SNP genotype file was phased with Beagle v5.153 and converted from the IUPAC ambiguity codes to a dominant genetic model, where ‘1’ represents the homozygous SNP states and ‘0’ represents the heterozygous sites. Each vector of genotype states for each marker was matched against the phenotype data for PLLL, PSLL, and PTD to perform iterative independent Kruskal–Wallis H-Tests with a custom loop created in R version 3.6.150. The output provided p values for each SNP marker across all three traits and each year of data collection to identify significantly associated SNPs. Highly reliable marker-trait associations were determined through the criteria of high statistical significance with p < 0.001, their detection in multiple years and significance in leaf and shoot susceptibility. The fire blight susceptibility values of SNPs satisfying these criteria were plotted to observe the effect the genotype state has on specific traits. To find patterns across wild and domesticated Malus species, the proportion and frequency of species that possessed each selected SNP state was analyzed. The LD relationship of each pairwise marker combination was calculated in order to assess the relation of selected SNPs to the neighboring markers. This was performed first with the PLINK v1.90b6 genomic software to generate .ped and .info file format54. The files were imported in the Haploview v4.2 software to visualize marker relationships and find haploblocks55.
Results
Genetic diversity in homologs of HIPM and DIPM
Nine homologs of HIPM and DIPM genes were identified in the GDDH13 apple genome sequence43 with sequence length ranging from 1.4 to 3.7 kilobases (Table 1). These genes were present across 8 chromosomes in the apple genome. Two duplicated gene copies were present for DIPM2, DIPM3, DIPM4, and HIPM1, whereas DIPM1 had a single copy present in the GDDH13 genome. It appeared that two copies of DIPM2 located on Chr13 likely evolved through tandem duplication events due to their close proximity. The duplicated copies from remaining HIPM and DIPM genes were distributed across different chromosomes that might have occurred through whole-genome duplication events.
To assess the genetic diversity across DIPM and HIPM genes, we used publicly available whole-genome sequences from 145 diverse Malus accessions (collection 1). A total of 677 SNP variants were identified across these genes. The least variable gene was DIPM4a with 18 variants while the highest, 189 variants, were found in HIPM1a (Table 1). Nucleotide diversity (π) varied from 6.02 × 10–03 to 2.66 × 10–03 for DIPM4a and DIPM2a, respectively, with an average π of 1.38 × 10–02. The gene annotation analysis identified 65 total variants as nonsynonymous mutations that can lead to amino acid changes in a protein. There were also 3 frameshift and 2 non-frameshift Indels present in HIPM1a, DIPM3a, and DIPM4b genes. Furthermore, inference of population genetic structure using principal component analysis (PCA) with 677 SNP variants did not reveal any distinct clusters across the 145 Malus accessions (Supplementary Figure S5A,B). Altogether, these results demonstrated the high level of genomic diversity present in the DIPM and HIPM genes in apples.
The lack of phenotypic information for accessions in this dataset hindered the possibility of making an association analysis between SNP markers and fire blight susceptibility. Thus, amplicon sequencing was employed to amplify and assess the genetic diversity in the HIPM and DIPM genes in 93 selected apple accessions (collection 2; Supplementary Table S1)44. The latter analysis identified a total of 87 SNP variants across 9 DIPM and HIPM genes ranging from 4 (HIPM1b) to 16 (DIPM2a) (Table 1). The average π was 7.96 × 10–03 in 9 genes that varied from 4.5 × 1003 (DIPM4a) to 1.8 × 1002 (HIPM1a). Moreover, annotation of gene sequences revealed that 19 SNP mutations lead to changes in amino acid sequences of proteins in the DIPM gene homologs. At least one amino acid-changing mutation was identified in each of the DIPM genes. For instance, a single nucleotide change from “G” to “A” (position = 379) in DIPM4a can change the amino acid glycine to serine (Supplementary Table S2). Similarly, three different SNP mutations in the DIPM4b gene can change amino acid sequence from valine to aspartic acid, phenylalanine to leucine, and lysine to arginine. We further used the DIPM-HIPM SNP dataset to infer population genetic structure across 93 diverse Malus accessions from Collection 21 using PCA. Two accessions were removed from the analysis because they had more than 50% missing data. PCA identified three groups in the 93 Malus accessions. A set of 7 accessions including GMAL 2590, GMAL 2366, GMAL 1527.f1, GMAL 3052.g1, ‘Prince George’, and ‘Glabrata’ showed close grouping along PC1 values less than − 3.0 (Supplementary Figure S5B). Another set of 9 GMAL and “Kola'' accessions form a dispersed group with PC1 values between − 1.0 and − 2.0 (Supplementary Figure S5B). Finally, the third and largest group consisted of 77 accessions with highly diverse genetic backgrounds (Supplementary Figure S5B; Supplementary Table S1).
Comparative variant analysis between collection 1 and collection 2 indicated higher genomic variation and amino acid-changing mutations across HIPM/DIPM genes from 145 accessions, probably due to near complete coverage of these gene sequences in the GDDH13 apple genome. However, thirteen annotated variants were shared between 145 NCBI and 93 AmpSeq samples, out of which only 2 on the gene “MD08G1095100” were nonsynonymous (Table 1).
Variation in fire blight susceptibility across diverse Malus accessions
Fire blight susceptibility in 93 Malus accessions was summarized as percentage of total infection as well as infection in the leaf and shoot (Supplementary Figure S2). The distributions of the leaf lengths and shoot lengths were normal across all accessions. The percent leaf lesion length (PLLL) was bimodal, centered around the extremes of the severity scale, the percent shoot lesion length (PSLL) was right skewed, and total lesion length measured as percent total distance (PTD) was also right skewed (Supplementary Figure S1). In 2018, the mean SL, LL, PSLL, PLLL, and PTD were 16.3 cm, 4.8 cm, 6.1%, 39.4%, and 12.1%, respectively. In 2019, the mean SL, LL, PSLL, PLLL, and PTD were 18.6 cm, 4.1 cm, 6.8%, 50.4%, and 13.7%, respectively. On the more resistant side of that range, there is a mix of accessions from M. baccata, and M. fusca as well as a few domesticated accessions such as ‘Koidu Rennett’. The highly susceptible accessions in the population included mostly M. domestica accessions such as ‘Idared’. The non-parametric Fligner-Killeen test for non-normal traits showed that the PLLL and PTD had homogenous variances across the accessions in the study (p value 2018/2019; PLLL: 0.8922, 0.1576, PTD: 0.1874, 0.5331). Overall, a wide range of responses to fire blight infection with varying levels of standard deviation were found across replicates for PLLL, PSLL, and PTD (Supplementary Figure S2A–C). Less susceptible accessions had relatively small standard deviations that increased as the values for PLLL and PTD increased consistently in both years (Supplementary Figure S2A,C).
The variation in PLLL, PSLL, and PTD during 2018 and 2019 was mainly determined by the genotypes (Table 2). For example, the ANOVA results showed that genotypic effects significantly influence fire blight susceptibility levels consistently for both leaf lesion (PLLL), shoot lesion (PSLL) and total lesion length in both years (Table 2). The k-mean clustering analysis of phenotypes showed larger ellipses for the susceptible class compared to the resistant class (Supplementary Figure S3). The clustering of genotypes based on the PLLL and PTD values identified three fire blight infection severity groups: susceptible, moderate, and resistant. The increase in fire blight severity of the susceptible group compared to the resistant group was 66.7% for PLLL and 27.2% for PTD in 2018. In 2019, the increase in fire blight severity of the susceptible group compared to the resistant group was 79.3% for PLLL and 36.3% for PTD.
Association between fire blight susceptibility and SNPs in HIPM and DIPM
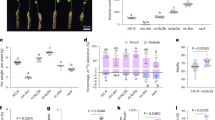
The Kruskal–Wallis H-test performed in R Version 3.6.250 identified 30 unique SNP markers that showed significant association to fire blight susceptibility at p < 0.05, 2 of which showed high significance at p < 0.001 (Fig. 1; Supplementary Table S3). There are 6 SNPs out of 30 significant SNPs that result into non-synonymous amino acid changes in HIPM and DIPM gene homologs. In addition, SNP markers showing significant associations with PTD and PLLL were identified in both 2018 and 2019. A SNP marker, S13_2611083, showed a significant association (p < 0.05) with PLLL, PTD in 2018 and PSLL in 2019 (p value = 0.046, 0.005, 0.017). A high level of significance (p < 0.001) was found for PLLL and PTD in 2019 (p value = 6.76e−05,1.66e−04) (Supplementary Table S3). This marker was present in the DIPM2b gene on chromosome 13. Another SNP marker S14_14033636, on the DIPM4b gene region on chromosome 14, showed significant association (p < 0.001) with all three traits in 2019 (Fig. 1).
Manhattan plots showing SNP markers significantly associated with fire blight susceptibility traits in 93 Malus accessions. Candidate gene association analysis was done using Kruskal Wallis H-Test for fire blight susceptibility data collected during 2018 and 2019. The x-axis represents the gene IDs and corresponding markers on HIPM/DIPM genes. The y-axis shows the − log10 (p values) of marker-trait associations for fire blight severity traits; PSLL (percent shoot lesion length), PLLL (percent leaf lesion length), and PTD (percent total distance). The PSLL, PLLL, and PTD traits are displayed as a circle, triangle, and square, respectively. Two red lines parallel to the x-axis indicate varying levels of alpha significance (alpha = 0.05, bottom; alpha = 0.001, top). Increased value of the − log10 (p value) indicates a higher level of significance for the marker-trait association. The left and right panels are derived from data collected in 2018 and 2019, respectively. The color of dot in each symbol corresponds to the line under SNP label.
According to haplotype analysis, the SNPs significantly associated with fire blight susceptibility were in low linkage disequilibrium (LD) with flanking markers (Supplementary Figure S4A,B). Boxplots of the homozygous and heterozygous genotypes at these SNP states showed clear phenotypic differences in their susceptibility levels (Fig. 2). The heterozygous state for S13_2611083 SNP marker had higher rates of fire blight severity and conferred 1.8 and 2.6-fold increases in susceptibility in 2018 for PLLL, and PTD, respectively (Fig. 2A). The homozygous allele state of S14_14033636 SNP marker was associated with higher fire blight susceptibility and showed 5.4, 2.5, and 3.7-fold increases in susceptibility in 2019 for PSLL, PLLL, and PTD, respectively (Fig. 2B). In 2019 for the same marker, there were 2.5, 2.2, and 2.6 fold increases in susceptibility for PSLL, PLLL, and PTD from changing the homozygous SNP to the heterozygous.
Boxplots of fire blight susceptibility at homozygous and heterozygous states of significantly associated SNP markers (A) S13_2611083 and (B) S14_14033636, respectively. The y-axis represents phenotypic values for percent leaf lesion length (PLLL), percent shoot lesion length (PSLL), and percent total distance (PTD). The “G” and “A” represent homozygous, and “K” and “R” represent heterozygous genotypic states for S13_2611083 and S14_14033636, respectively, on the x-axis. The table indicates allele frequency of homozygous and heterozygous SNP states across studied genotypes.
We further observed additive allelic effects on the fire blight susceptibility from the most significant SNPs across HIPM and DIPM genes. For example, accessions possessing alleles linked with reduced susceptibility from both S13_2611083 and S14_14033636 markers showed an additive phenotypic response of significantly less fire blight susceptibility for PLLL (p value = 2018: 0.011, 2019: 0.0084) and PTD (p value = 2018: 0.019, 2019: 0.0011) and vice-versa (Fig. 3). The differences for PSLL were only significant in 2019 between the most susceptible class ‘A + K’ and the two less susceptible classes ‘R + K’, and ‘R + G’ (p value = 0.007,0.02; respectively) (Fig. 3).
Pairwise Kruskal–Wallis H-tests to measure the additive effects of S13_2611083 and S14_14033636 significant SNP (single nucleotide polymorphism) markers in years (A) 2018, and (B) 2019. The x-axis represents one of the four possible combinations of homozygous and heterozygous genotype states from markers S13_2611083 (G or K) and S14_14033636 (A or R). The y-axis represents the fire blight susceptibility response in accessions carrying specific allele combinations. Fire blight susceptibility was measured as PLLL (percent leaf lesion length), PSLL (percent shoot lesion length), and PTD (percent total distance). The numbers on top of brackets for each subplot represent the p value of significance for pairwise comparison of each allelic combination for a specific trait and year.
The distribution of marker alleles across Malus species showed that the allelic states linked with higher susceptibility levels were mostly present in M. domestica accessions (Fig. 4A,B). In 2018, the average increase of susceptibility in domesticated accessions as opposed to wild accessions were 2.13, 6.12, and 5.22% for PLLL, PSLL, and PTD, respectively. In 2019, the average increases of susceptibility in domesticated accessions were 13.67, 6.68, and 5.5% for PLLL, PSLL, and PTD, respectively. SNP alleles with reduced levels of susceptibility mainly occurred in wild Malus species, whereas the proportion of higher susceptibility SNPs were found in M. domestica (Fig. 4A,B).
Horizontal barplots showing the frequency and proportion of Malus species that possess more fire blight susceptible heterozygous ‘K’ or ‘A’ or the less susceptible homozygous ‘G’ or ‘R’ SNP genotype at SNP marker (A) S13_2661083 and (B) S14_14033636, respectively. The y-axis indicates the 14 Malus species with either of the two SNP genotypes. The frequency is defined by the number of occurrences of a SNP across all the genotypes. The proportion is calculated by the frequency of a SNP divided by the total number of SNP, multiplied by 100.
Discussion
Genomic and genetic characterization of the S genes, which are responsible for successful pathogen infection, is a logical approach to achieve sustainable disease tolerance in improved cultivars56,57. Natural genetic variation in these genes can be tapped to develop cultivars with decreased fire blight susceptibility, a major goal of apple germplasm improvement efforts worldwide. The susceptibility genes, HIPMs and DIPMs, have been identified to regulate susceptibility and improved tolerance against fire blight in commercial apple cultivars35,38,40,58. We have found a great number of genomic variations in the 9 HIPM and DIPM genes across 145 diverse Malus accessions. We also found 30 SNPs with significant (p < 0.05) associations to fire blight susceptibility, two of which had strong associations in two consecutive years of fire blight infection. A majority of the marker allelic states related to higher levels of susceptibility in these trait-associated markers were found in M. domestica, which is consistent with the current understanding that domesticated cultivars are generally susceptible to fire blight7. A higher proportion of species with the less susceptible allele were seen in wild species, with a shift to 50:50 in Malus hybrids. This is a possible indication of an inheritance pattern where a new mutation is introduced in a population and the more susceptible alleles are maintained and proliferated by selection during apple domestication. These susceptibility alleles were likely maintained in the domesticated germplasm due to fewer instances of cross-hybridization with wild apple species, to avoid unnecessary linkage drag of undesirable traits affecting fruit quality in apple cultivars.
The SNP marker S13_2611083 on DIPM2b showed significant association (p < 0.05) with PLLL and PTD in 2018 and (p < 0.001) in 2019. Robustness and stability of this SNP marker across two years makes it a reliable marker to screen for fire blight susceptibility and for use in marker-assisted breeding. Similarly, the strong association (p < 0.001) of SNP marker S14_14033636 on the DIPM4b gene with PLLL and PTD indicates its potential use in marker-assisted selection to decrease fire blight susceptibility. The latter finding is consistent with previous work in which knocking out the MdDIPM4 gene via CRISPR/Cas9 led to a 50% reduction in fire blight symptoms on notoriously susceptible ‘Gala’ and ‘Golden Delicious' cultivars40. These results also highlight utilization of candidate-gene based association studies to rapidly identify molecular markers linked to traits of interest for progeny and parent selection in breeding programs59. Breeding from a perspective of susceptibility rather than resistance can help resolve issues in tree fruit crops where linkage drag and cross incompatibility between Malus species and cultivated apples makes introgression of resistance alleles not feasible or very time consuming in many cases11. Hence, S-gene breeding opens the opportunity for breeders to more effectively improve fire blight resiliency of high value apple cultivars while not compromising on critical consumer focused traits.
Our results suggest that additive effects from multiple small effect loci in HIPM and DIPM genes contribute towards overall variation observed in disease susceptibility of domesticated apples. For example, combinations of positive alleles of S13_2611083 and S14_14033636 SNPs from two different loci have a stronger effect on fire blight susceptibility measures (i.e., PLLL and PTD). This is in contrast with the effect of major resistances contributed by wild apple germplasm, where a single gene or locus determined the fire blight resistance response without interacting with other loci in the genome16,60,61,62. The breakdown of single gene resistance/s by novel fire blight strains is highly likely, as shown for the breakdown of Robusta 5 (MR5) fire blight resistance by a deletion in the avrRpt2 effector of Ea1189 E. amylovora strain20. Similarly, different strains of E. amylovora have been identified as virulent and avirulent on apple cultivars engineered for MR5 resistance21. These studies indicate that introgression of single gene resistances into commercial germplasm lie at a risk of breakdown by rapidly evolving pathogen strains. Emergence of a virulent strain for the particular resistance gene poses significant threat to breeding and selection efforts based on few major genes. Alternatively, additive alleles from different pathways can help achieve durable reduced susceptibility in a cultivar. For instance, selection of appropriate allelic combinations from HIPM/DIPM genes can lower successful pathogen infection, leading to reduced fire blight susceptibility. The root system can also influence fire blight susceptibility levels through co-regulated gene expression patterns and system-level interactions between carbohydrate and defense pathways63. Therefore, allelic combinations for the optimal root system can be used to breed rootstocks to further reduce fire blight susceptibility of grafted scions. These results depict a few examples related to disease susceptibility regulation, but apparently there are still several unexplored routes associated with disease susceptibility in the domesticated apples. Nonetheless, the presence of additive gene action from multiple pathways that are associated with disease susceptibility can make it harder for new pathogens to completely overcome the reduced disease susceptibility, which could be feasible to improve disease tolerance in apple breeding programs56.
The results presented in this study also represent strain-specific differences in host plant responses to fire blight as observed in the identification of different significant SNPs between 2018 and 2019. A mixture of three strains (Ea273, E4001a, E2002a) was used in 2018 while only E2002a was used in 2019 to inoculate the plants. The pure inoculum of the highly virulent E2002a strain generally showed higher disease incidence in 2019 for PLLL and PTD (Figure S2). In the mixed inoculum of 2018 infections, E2002a was present in lower concentrations and probably had more competition for host resources with Ea273 and E4001a. Furthermore, E2002a is a native North American strain, first discovered in Ontario, Canada off the “Jonathan” cultivar64,65. This strain likely had higher co-evolutionary compatibility with the North American domesticated accessions in this study. The chances of host–pathogen compatibility are mainly determined by the profile of effector proteins in individual E. amylovora strains that are responsible for fire blight pathogenicity and triggering of the hypersensitive response in Malus6,66. However, the strains used in this study exhibit high genome similarity49 and their considerably different phenotypic and genetic responses in host plants can lay out further investigations of Malus-Erwinia interactions.
In summary, a great genomic diversity was found in HIPM and DIPM genes in a wide set of Malus accessions and a candidate gene-based approach identified markers that were strongly associated with fire blight susceptibility. The validation of these markers could provide opportunities to use them in MAS to breed apple cultivars with reduced fire blight susceptibility. Moreover, characterization of S-genes can provide an alternative to improve fire blight resistance, either by breeding or genome-editing to avoid unnecessary risks associated with compromised fruit quality and breakdown of major resistance genes from wild Malus species.
Change history
27 January 2021
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Flachowsky, H. et al. Application of a high-speed breeding technology to apple (Malus×domestica) based on transgenic early flowering plants and marker-assisted selection. New Phytol. https://doi.org/10.1111/j.1469-8137.2011.03813.x (2011).
Grattapaglia, D. & Resende, M. D. V. Genomic selection in forest tree breeding. Tree Genet. Genomes https://doi.org/10.1007/s11295-010-0328-4 (2011).
Kumar, S., Bink, M. C. A. M., Volz, R. K., Bus, V. G. M. & Chagné, D. Towards genomic selection in apple (Malus × domestica Borkh.) breeding programmes: prospects, challenges and strategies. Tree Genet. Genomes https://doi.org/10.1007/s11295-011-0425-z (2012).
Resende, M. D. V. et al. Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol. https://doi.org/10.1111/j.1469-8137.2011.04038.x (2012).
Ghosh, S. et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. https://doi.org/10.1038/s41596-018-0072-z (2018).
Khan, M. A., Zhao, Y. & Korban, S. S. Molecular mechanisms of pathogenesis and resistance to the bacterial pathogen Erwinia amylovora, causal agent of fire blight disease in rosaceae. Plant Mol. Biol. Rep. https://doi.org/10.1007/s11105-011-0334-1 (2012).
Norelli, J. L., Jones, A. L. & Aldwinckle, H. S. Fire blight management in the twenty-first century: using new technologies that enhance host resistance in apple. Plant Dis. https://doi.org/10.1094/PDIS.2003.87.7.756 (2003).
Luo, F., Evans, K., Norelli, J. L., Zhang, Z. & Peace, C. Prospects for achieving durable disease resistance with elite fruit quality in apple breeding. Tree Genet. Genomes https://doi.org/10.1007/s11295-020-1414-x (2020).
Muñoz-Sanz, J. V., Zuriaga, E., Cruz-García, F., McClure, B. & Romero, C. Self-(in)compatibility systems: target traits for crop-production, plant breeding, and biotechnology. Front. Plant Sci. https://doi.org/10.3389/fpls.2020.00195 (2020).
Longhi, S. et al. A candidate gene based approach validates Md-PG1 as the main responsible for a QTL impacting fruit texture in apple (Malus × domestica Borkh). BMC Plant Biol https://doi.org/10.1186/1471-2229-13-37 (2013).
Malnoy, M. et al. Fire blight: applied genomic insights of the pathogen and host. Annu. Rev. Phytopathol. https://doi.org/10.1146/annurev-phyto-081211-172931 (2012).
Busdieker-Jesse, N. L., Nogueira, L., Onal, H. & Bullock, D. S. The economic impact of new technology adoption on the U.S. apple industry. J. Agric. Resour. Econ. https://doi.org/10.22004/ag.econ.246254 (2016).
Beckerman, J. L. & Sundin, G. W. Scab and fire blight of apple: issues in integrated pest management. Hortic. Rev. https://doi.org/10.1002/9781119281269.ch8 (2016).
Peil A, Hanke M V., Flachowsky H et al. Confirmation of the fire blight QTL of Malus × robusta 5 on linkage group 3. In: Acta Horticulturae (2008).
Durel, C. E., Denancé, C. & Brisset, M. N. Two distinct major QTL for resistance to fire blight co-localize on linkage group 12 in apple genotypes ‘Evereste’ and Malus floribunda clone 821. Genome https://doi.org/10.1139/G08-111 (2009).
Emeriewen, O. et al. Identification of a major quantitative trait locus for resistance to fire blight in the wild apple species Malus fusca. Mol. Breed. https://doi.org/10.1007/s11032-014-0043-1 (2014).
Calenge, F. et al. Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theor. Appl. Genet. https://doi.org/10.1007/s00122-005-2002-z (2005).
Khan, M. A., Duffy, B., Gessler, C. & Patocchi, A. QTL mapping of fire blight resistance in apple. Mol. Breed. https://doi.org/10.1007/s11032-006-9000-y (2006).
Khan, M. A., Zhao, Y. F. & Korban, S. S. Identification of genetic loci associated with fire blight resistance in Malus through combined use of QTL and association mapping. Physiol. Plant https://doi.org/10.1111/ppl.12068 (2013).
Vogt, I. et al. Gene-for-gene relationship in the host-pathogen system Malus × robusta 5-Erwinia amylovora. New Phytol. https://doi.org/10.1111/nph.12094 (2013).
Broggini, G. A. L. et al. Engineering fire blight resistance into the apple cultivar ‘Gala’ using the FB_MR5 CC-NBS-LRR resistance gene of Malus × robusta 5. Plant Biotechnol. J. https://doi.org/10.1111/pbi.12177 (2014).
van Schie, C. C. N. & Takken, F. L. W. Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. https://doi.org/10.1146/annurev-phyto-102313-045854 (2014).
Candresse, T., Le Gall, O., Maisonneuve, B., German-Retana, S. & Redondo, E. The use of green fluorescent protein-tagged recombinant viruses to test Lettuce mosaic virus resistance in lettuce. Phytopathology https://doi.org/10.1094/PHYTO.2002.92.2.169 (2002).
Nicaise, V. et al. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol https://doi.org/10.1104/pp.102.017855 (2003).
Gao, Z. et al. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor elF4E in cell-to-cell trafficking. Plant J https://doi.org/10.1111/j.1365-313X.2004.02215.x (2004).
Gao, Z., Eyers, S., Thomas, C., Ellis, N. & Maule, A. Identification of markers tightly linked to sbm recessive genes for resistance to Pea seed-borne mosaic virus. Theor. Appl. Genet. https://doi.org/10.1007/s00122-004-1652-6 (2004).
Kang, B. C., Yeam, I., Frantz, J. D., Murphy, J. F. & Jahn, M. M. The pvr1 locus in Capsicum encodes a translation initiation factor elF4E that interacts with Tobacco etch virus VPg. Plant J. https://doi.org/10.1111/j.1365-313X.2005.02381.x (2005).
Morales, M. et al. A physical map covering the nsv locus that confers resistance to Melon necrotic spot virus in melon (Cucumis melo L.). Theor. Appl. Genet. https://doi.org/10.1007/s00122-005-0019-y (2005).
Nieto, C. et al. An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J. https://doi.org/10.1111/j.1365-313X.2006.02885.x (2006).
Iyer-Pascuzzi, A. S. & McCouch, S. R. Recessive resistance genes and the Oryza sativa—Xanthomonas oryzae pv. oryzae pathosystem. Mol. Plant Microbe Interact. https://doi.org/10.1094/MPMI-20-7-0731 (2007).
Rakotomalala, M. et al. Resistance to rice yellow mottle virus in rice germplasm in Madagascar. Eur. J. Plant Pathol. https://doi.org/10.1007/s10658-008-9282-5 (2008).
Tyrka, M., Perovic, D., Wardyńska, A. & Ordon, F. A new diagnostic SSR marker for selection of the Rym4/Rym5 locus in barley breeding. J. Appl. Genet https://doi.org/10.1007/BF03195605 (2008).
Jia, H. et al. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. https://doi.org/10.1111/pbi.12677 (2017).
Borejsza-Wysocka, E. E. et al. The fire blight resistance of apple clones in which DspE-interacting proteins are silenced. In X International Workshop on Fire Blight. Acta Hort. 704, 509–513 (2006).
Malnoy, M. et al. Silencing of HIPM, the apple protein that interacts with HrpN of Erwinia amylovora. Acta Hortic. https://doi.org/10.17660/ActaHortic.2008.793.38 (2008).
Oh, C. S. & Beer, S. V. AtHIPM, an ortholog of the apple HrpN-interacting protein, is a negative regulator of plant growth and mediates the growth-enhancing effect of HrpN in arabidopsis. Plant Physiol. https://doi.org/10.1104/pp.107.103432 (2007).
Bocsanczy, A. M., Nissinen, R. M., Oh, C. S. & Beer, S. V. HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol. Plant Pathol. https://doi.org/10.1111/j.1364-3703.2008.00471.x (2008).
Campa, M. et al. HIPM is a susceptibility gene of Malus spp.: reduced expression reduces susceptibility to Erwinia amylovora. Mol. Plant Microbe Interact. https://doi.org/10.1094/MPMI-05-18-0120-R (2019).
Meng, X., Bonasera, J. M., Kim, J. F., Nissinen, R. M. & Beer, S. V. Apple proteins that interact with DspA/E, a pathogenicity effector of Erwinia amylovora, the fire blight pathogen. Mol. Plant Microbe Interact. https://doi.org/10.1094/MPMI-19-0053 (2006).
Pompili, V., Dalla Costa, L., Piazza, S., Pindo, M. & Malnoy, M. Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. https://doi.org/10.1111/pbi.13253 (2020).
Duan, N. et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. https://doi.org/10.1038/s41467-017-00336-7 (2017).
Bianco, L. et al. Development and validation of the Axiom Apple480K SNP genotyping array. Plant J. https://doi.org/10.1111/tpj.13145 (2016).
Daccord, N. et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. https://doi.org/10.1038/ng.3886 (2017).
Yang, S. et al. A next-generation marker genotyping platform (AmpSeq) in heterozygous crops: a case study for marker-assisted selection in grapevine. Hortic. Res. https://doi.org/10.1038/hortres.2016.2 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics https://doi.org/10.1093/bioinformatics/btu170 (2014).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. https://doi.org/10.1093/nar/gkq603 (2010).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics https://doi.org/10.1093/bioinformatics/btr330 (2011).
Bradbury, P. J. et al. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics https://doi.org/10.1093/bioinformatics/btm308 (2007).
Singh, J. & Khan, A. Distinct patterns of natural selection determine sub-population structure in the fire blight pathogen, Erwinia amylovora. Sci. Rep. https://doi.org/10.1038/s41598-019-50589-z (2019).
R Core Team. R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. (R Foundation for Statistical Computing, 2020).
Desnoues, E. et al. Identification of novel strain-specific and environment-dependent minor QTLs linked to fire blight resistance in apples. Plant Mol. Biol. Rep. https://doi.org/10.1007/s11105-018-1076-0 (2018).
Charrad, M., Ghazzali, N., Boiteau, V. & Niknafs, A. Nbclust: An R package for determining the relevant number of clusters in a data set. J. Stat. Softw. https://doi.org/10.18637/jss.v061.i06 (2014).
Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. https://doi.org/10.1086/521987 (2007).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. https://doi.org/10.1086/519795 (2007).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics https://doi.org/10.1093/bioinformatics/bth457 (2005).
Pavan, S., Jacobsen, E., Visser, R. G. F. & Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. https://doi.org/10.1007/s11032-009-9323-6 (2010).
Zaidi, S. S., Mukhtar, M. S. & Mansoor, S. Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2018.04.005 (2018).
Borejsza-Wysocka, E. E. et al. The fire blight resistance of apple clones in which DspE-interacting proteins are silenced. Acta Hortic. https://doi.org/10.17660/actahortic.2006.704.80 (2006).
Udoh, L. I. et al. Candidate gene sequencing and validation of SNP markers linked to carotenoid content in cassava (Manihot esculenta Crantz). Mol. Breed. https://doi.org/10.1007/s11032-017-0718-5 (2017).
Parravicini, G. et al. Identification of serine/threonine kinase and nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes in the fire blight resistance quantitative trait locus of apple cultivar ‘Evereste’. Mol. Plant Pathol. https://doi.org/10.1111/j.1364-3703.2010.00690.x (2011).
Fahrentrapp, J. et al. A candidate gene for fire blight resistance in Malus × robusta 5 is coding for a CC-NBS-LRR. Tree Genet. Genomes https://doi.org/10.1007/s11295-012-0550-3 (2013).
Emeriewen, O. F. et al. Fire blight resistance of Malus ×arnoldiana is controlled by a quantitative trait locus located at the distal end of linkage group 12. Eur. J. Plant Pathol. https://doi.org/10.1007/s10658-017-1152-6 (2017).
Singh, J. et al. Root system traits impact early fire blight susceptibility in apple (Malus × domestica). BMC Plant Biol. https://doi.org/10.1186/s12870-019-2202-3 (2019).
Quamme, H. A. & Bonn, W. G. Virulence of Erwinia amylovora and its influence on the determination of fire blight resistance of pear cultivars and seedlings. Can. J. Plant Pathol. https://doi.org/10.1080/07060668109501345 (1981).
Norelli, J. L. Differential host × pathogen interactions among cultivars of apple and strains of Erwinia amylovora. Phytopathology https://doi.org/10.1094/phyto-74-136 (1984).
Emeriewen, O. F., Wöhner, T., Flachowsky, H. & Peil, A. Malus hosts–Erwinia amylovora interactions: strain pathogenicity and resistance mechanisms. Front. Plant Sci. https://doi.org/10.3389/fpls.2019.00551 (2019).
Acknowledgements
This research was financially supported by the New York State Apple and Research Development Program (ARDP). We acknowledge Della Cobb-Smith (Cornell University) for help during the evaluation of the fire blight data in the greenhouse.
Author information
Authors and Affiliations
Contributions
A.K. and M.M. conceived, designed and supervised the research. R.T., and K.J.P.S. conducted the fire blight evaluation in the greenhouse. R.T., V.P., J.S., and D.M. performed the amplicon sequencing, bioinformatics, genetic and statistical analysis. R.T. wrote the first draft of the manuscript and all authors contributed towards improving and reviewing the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tegtmeier, R., Pompili, V., Singh, J. et al. Candidate gene mapping identifies genomic variations in the fire blight susceptibility genes HIPM and DIPM across the Malus germplasm. Sci Rep 10, 16317 (2020). https://doi.org/10.1038/s41598-020-73284-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-73284-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.