« Prev Next »
It is estimated that the human genome encodes approximately 25,000 genes, about the same number as that for corn and nearly twice as many as that for the common fruit fly. Even more interesting is the fact that those 25,000 genes are encoded in about 1.5% of the genome. So, what exactly does the other 98.5% of our DNA do? While many mysteries remain about what all of that extra sequence is for, we know that it does contain complex instructions that direct the intricate turning on and off of gene transcription.
Eukaryotes Require Complex Controls Over Gene Expression
While basic similarities in gene transcription exist between prokaryotes and eukaryotes—including the fact that RNA polymerase binds upstream of the gene on its promoter to initiate the process of transcription—multicellular eukaryotes control cell differentiation through more complex and precise temporal and spatial regulation of gene expression.
Multicellular eukaryotes have a much larger genome than prokaryotes, which is organized into multiple chromosomes with greater sequence complexity. Many eukaryotic species carry genes with the same sequences as other plants and animals. In addition, the same DNA sequences (though not the same proteins) are found within all of an organism's diploid, nucleated cells, even though these cells form tissues with drastically different appearances, properties, and functions. Why then, is there such great variation among and within such organisms? Quite simply, the way in which different genes are turned on and off in specific cells generates the variety we observe in nature. In other words, specific functions of different cell types are generated through differential gene regulation.
Of course, higher eukaryotes still respond to environmental signals by regulating their genes. But there is an additional layer of regulation that results from cell-to-cell interactions within the organism that orchestrate development. Specifically, gene expression is controlled on two levels. First, transcription is controlled by limiting the amount of mRNA that is produced from a particular gene. The second level of control is through post-transcriptional events that regulate the translation of mRNA into proteins. Even after a protein is made, post-translational modifications can affect its activity.
Transcriptional Regulation in Eukaryotes
Regulation of transcription in eukaryotes is a result of the combined effects of structural properties (how DNA is "packaged") and the interactions of proteins called transcription factors. The most important structural difference between eukaryotic and prokaryotic DNA is the formation of chromatin in eukaryotes. Chromatin results in the different transcriptional "ground states" of prokaryotes and eukaryotes (Table 1).
Table 1: Overview of Differences Between Prokaryotic and Eukaryotic Gene Expression and Regulation
| Prokaryotes | Eukaryotes | |
| Structure of genome | Single, generally circular genome sometimes accompanied by smaller pieces of accessory DNA, like plasmids | Genome found in chromosomes; nucleosome structure limits DNA accessibility |
| Size of genome | Relatively small | Relatively large |
| Location of gene transcription and translation | Coupled; no nucleoid envelope barrier because of prokaryotic cell structure | Nuclear transcription and cytoplasmic translation |
| Gene clustering | Operons where genes with similar function are grouped together | Operons generally not found in eukaryotes; each gene has its own promoter element and enhancer element(s) |
| Default state of transcription | On | Off |
| DNA structure | Highly supercoiled DNA with some associated proteins | Highly supercoiled chromatin associated with histones in nucleosomes |
Transcription Factors and Combinatorial Control
Sequence-specific transcription factors are considered the most important and diverse mechanisms of gene regulation in both prokaryotic and eukaryotic cells (Pulverer, 2005). In eukaryotes, regulation of gene expression by transcription factors is said to be combinatorial, in that it requires the coordinated interactions of multiple proteins (in contrast to prokaryotes, in which a single protein is usually all that is required).
Many genes, known as housekeeping genes, are needed by almost every type of cell and appear to be unregulated or constitutive. But at the core of cellular differentiation, manifested in the variety of cell types observed in different organisms, is the regulation of gene expression in a tissue-specific manner. The same genome is responsible for making the entire cadre of cell types, each of which has its own function—for example, red blood cells exchange oxygen, muscle cells expand and contract, and cells in the immune system recognize pathogens. Genes that regulate cell identity are turned on under very specific temporal, spatial, and environmental conditions to ensure that a cell is able to perform its designated function.
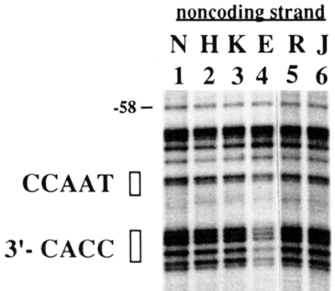
Take the example of the gene for beta globin, a protein used in red blood cells for oxygen exchange. Every cell in the human body contains the beta globin gene and the corresponding upstream regulatory sequences that regulate expression, but no cell type other than red blood cells expresses beta globin. Scientists can use a technique called DNA footprinting to map where transcription factors bind to specific regulatory sequences. When Reddy et al. examined the beta globin promoter in different cell types, they found that the transcription factors that could bind to the promoter sequences required for beta globin expression were expressed only in erythroblasts (immature adult red blood cells). (See Figure 1). The two consensus sequences in the beta globin promoter known for binding transcription factors, CCAAT and CACC, were protected in the erythroid cells (E), but not the other cell types (Reddy et al., 1994).
The "Ground State" of DNA Expression
RNA polymerase in prokaryotes can access almost any promoter in a DNA strand without the presence of activators or repressors. Thus, the "ground state" of DNA expression in prokaryotes is said to be nonrestrictive, or "on." In eukaryotes, however, the ground state of expression is restrictive in that, although strong promoters might be present, they are inactive in the absence of some sort of recruitment to the promoter by transcription factors. For instance, RNA polymerase II, which transcribes mRNA, cannot bind to promoters in eukaryotic DNA without the help of transcription factors (Struhl, 1999). In many eukaryotic organisms, the promoter contains a conserved gene sequence called the TATA box. Various other consensus sequences also exist and are recognized by the different TF families. Transcription is initiated when one TF binds to one of these promoter sequences, initiating a series of interactions between multiple proteins (activators, regulators, and repressors) at the same site, or other promoter, regulator, and enhancer sequences. Ultimately, a transcription complex is formed at the promoter that facilitates binding and transcription by RNA polymerase.
As in prokaryotes, eukaryotic repressor molecules can sometimes bind to silencer elements in the vicinity of a gene and inhibit the binding, assembly, or activity of the transcription complex, thus turning off expression of a gene. Positive regulation by TFs that are activators is common in eukaryotes. Considering the restrictive transcriptional ground state, it is logical that positive regulation is the predominant form of control in all systems characterized to date. Many activating TFs are generally bound to DNA until removed by a signal molecule, while others might only bind to DNA once influenced by a signal molecule. The binding of one type of TF can influence the binding of others, as well. Thus, gene expression in eukaryotes is highly variable, depending on the type of activators involved and what signals are present to control binding.
The Role of Chromatin
Even when transcription factors are present in a cell, transcription does not always occur, because often the TFs cannot reach their target sequences. The association of the DNA molecule with proteins is the first step in its silencing. The associated DNA and histone proteins are collectively called chromatin; the complex is tightly bonded by attraction of the negatively charged DNA to the positively charged histones (Table 1). The state of chromatin can limit access of transcription factors and RNA polymerase to DNA promoters, contributing to the restrictive ground state of gene expression. In order for gene transcription to occur, the chromatin structure must be unwound.
Chromatin structure contributes to the varying levels of complexity in gene regulation. It allows simultaneous regulation of functionally or structurally related genes that tend to be present in widely spaced clusters or domains on eukaryotic DNA (Sproul et al., 2005). Interactions of chromatin with activators and repressors can result in domains of chromatin that are open, closed, or poised for activation. Chromatin domains have various sizes and different extents of stability. These variations allow for phenomena found solely in eukaryotes, such as transcription at various stages of development and epigenetic memory throughout cell division cycles. They also allow for the maintenance of differentiated cellular states, which is crucial to the survival of multicellular organisms (Struhl, 1999).
Multiple Interactions Provide Synchronous Control
As you have seen, the state of chromatin structure at a specific region in eukaryotic DNA, along with the presence of specific transcription factors, works to regulate gene expression in eukaryotes. However, this complex interplay between proteins that serve as transcriptional activators or repressors and accessibility to the regulatory sequence is still just part of the story. Epigenetic mechanisms, including DNA methylation and imprinting, noncoding RNA, post-translational modifications, and other mechanisms, further enrich the cellular portfolio of gene expression control activities.
References and Recommended Reading
Pulverer, B. Sequence-specific DNA-binding transcription factors. Nature Milestones (2005) doi: 10.1038/nrm1800 (link to article)
Reddy, P. M., Stamatoyannopoulos, G., Papayannopoulou, T., & Shen, C. K. Genomic footprinting and sequencing of human beta-globin locus: Tissue specificity and cell line artifact. Journal of Biological Chemistry 269, 8287–8295 (1994)
Remenyi, A., Scholer, H., & Wilmanns, M. Combinatorial control of gene expression. Nature Structural and Molecular Biology 11, 812–815 (2004) doi:10.1038/nsmb820 (link to article)
Struhl, K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98, 1–4 (1999)
Sproul, D., Gilbert, N., & Bickmore, W. The role of chromatin structure in regulating the expression of clustered genes. Nature Reviews Genetics 6, 775–781 (2005) doi:10.1038/nrg1688 (link to article)



 Figure 1: DNA footprinting reveals transcription factor specificity in different cell types
Figure 1: DNA footprinting reveals transcription factor specificity in different cell types


























