« Prev Next »
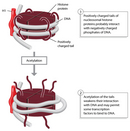
Within cells, chromosomal DNA associates with histones, forming an organized complex known as chromatin. Chromatin enables DNA to be packaged into a smaller volume so that it fits compactly within a cell's nucleus, and it also helps regulate gene expression. Specifically, compaction of the genome in the form of chromatin limits genes' accessibility to transcription factors. Thus, in order for gene expression to occur, changes in chromatin structure, called chromatin remodeling, must take place. These changes are brought about primarily by biochemical modifications to histones, including methylation, acetylation, and phosphorylation. Remodeling ultimately results in altered accessibility of transcription factors to regulatory DNA. Thus, understanding how chromatin remodeling is detected is a fundamental lesson in the study of gene regulation.
Detection of Chromatin Remodeling
As previously mentioned, chromatin is remodeled (and thus, nucleosome structure is altered) primarily through posttranslational modifications of histone proteins, which actually change the conformation of nearby DNA. For instance, histone acetylation reduces the positive charge of the histone proteins, thereby reducing their affinity for DNA and causing the chromatin to open (Figure 2). This in turn permits some transcription factors to bind to the DNA. Indeed, the "open" or "closed" state of the chromatin near a particular gene can be revealed by examining DNA sensitivity to the enzyme DNAse I by way of a procedure known as a DNAse sensitivity assay. This technique is based on the fact that DNAse I degrades open DNA more quickly than closed DNA. Not only can this type of assay provide information about whether gene expression changes are accompanied by chromatin remodeling, but it can also show where remodeling is taking place.
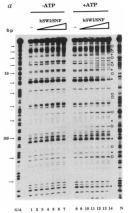
The hSWI/SNF Complex Leads to Increased DNAse Sensitivity
To better understand the exact role of the hSWI/SNF complex in chromatin remodeling, scientists conducted an experiment in which they exposed chromatin to increasing concentrations of this complex (Figure 3). As the concentration of the hSWI/SNF complex increased (indicated by the triangles along the top of the figure), so did the chromatin's level of DNAse sensitivity; by extension, this meant that greater levels of change in chromatin structure were associated with increasing concentrations of the hSWI/SNF complex. Indeed, changes in DNAse sensitivity can be seen in the gel in Figure 3. Gel lanes marked with an open circle indicate increased cleavage or sensitivity, while those marked with a closed circle indicate decreased sensitivity.
Importantly, during this experiment, many of the changes in DNAse sensitivity were observed only in the presence of ATP (compare the +ATP lanes with the -ATP lanes). These observations strongly suggest that hSWI/SNF-mediated chromatin remodeling is ATP-dependent. Because ATP is an energy source within cells, dependence on ATP indicates that remodeling is not likely to be a random or stochastic event. Rather, these observations strongly suggest that chromatin remodeling is a regulated process-one that has important implications for gene expression (Kwon et al., 1994). Thus, while chromatin was originally thought to play a simple structural role in cells, findings such as this have demonstrated the importance of chromatin in dictating changes in gene expression.
References and Recommended Reading
Kwon, H., et
al. Nucleosome disruption and enhancement of activator binding by a human
SW1/SNF complex. Nature 370, 477–481 (1994) (link to article)



 Figure 1
Figure 1