Key Points
-
Gene annotation is one of the core mechanisms through which we decipher the information that is contained in genome sequences.
-
Gene annotation is complicated by the existence of 'transcriptional complexity', which includes extensive alternative splicing and transcriptional events outside of protein-coding genes.
-
The annotation strategy for a given genome will depend on what it is hoped to achieve, as well as the resources available.
-
The availability of next-generation data sets has transformed gene annotation pipelines in recent years, although their incorporation is rarely straightforward.
-
Even human gene annotation is far from complete: transcripts are missing and existing models are truncated. Most importantly, 'functional annotation' — the description of what transcripts actually do — remains far from comprehensive.
-
Efforts are now under way to integrate gene annotation pipelines with projects that seek to describe regulatory sequences, such as promoter and enhancer elements.
-
Gene annotation is producing increasingly complex resources. This can present a challenge to usability, most notably in a clinical context, and annotation projects must find ways to resolve such problems.
Abstract
A genome sequence is worthless if it cannot be deciphered; therefore, efforts to describe — or 'annotate' — genes began as soon as DNA sequences became available. Whereas early work focused on individual protein-coding genes, the modern genomic ocean is a complex maelstrom of alternative splicing, non-coding transcription and pseudogenes. Scientists — from clinicians to evolutionary biologists — need to navigate these waters, and this has led to the design of high-throughput, computationally driven annotation projects. The catalogues that are being produced are key resources for genome exploration, especially as they become integrated with expression, epigenomic and variation data sets. Their creation, however, remains challenging.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gerstein, M. B. et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 17, 669–681 (2007). This influential article attempts to rationalize a modern description of the gene in the context of transcriptional complexity.
Harrow, J. et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). This provides a detailed description of the GENCODE annotation pipeline.
Kim, V. N., Han, J. & Siomi, M. C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 (2009).
Andersson, L. et al. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 16, 57 (2015).
O'Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). This is an excellent starting point for exploring the NCBI annotation resources.
McGarvey, K. M. et al. Mouse genome annotation by the RefSeq project. Mamm. Genome 26, 379–390 (2015).
Mudge, J. M. & Harrow, J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome 26, 366–378 (2015).
Berardini, T. Z. et al. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485 (2015).
Howe, K. L. et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 44, D774–D780 (2016).
Attrill, H. et al. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44, D786–D792 (2016).
Elsik, C. G. et al. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15, 86 (2014).
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016). This provides a detailed description and comparison of various RNA-seq analytical pipelines.
Boutet, E. et al. UniProtKB/Swiss-prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374, 23–54 (2016). The UniProt and Swiss-Prot resources are outlined here.
Stanke, M. & Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 (Suppl. 2), ii215–ii225 (2003).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997).
Yandell, M. & Ence, D. A beginner's guide to eukaryotic genome annotation. Nat. Rev. Genet. 13, 329–342 (2012).
Gray, K. A., Yates, B., Seal, R. L., Wright, M. W. & Bruford, E. A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 43, D1079–D1085 (2015).
Guigo, R. et al. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 7, S2 (2006).
Zhang, G. et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320 (2014).
Eory, L. et al. Avianbase: a community resource for bird genomics. Genome Biol. 16, 21 (2015).
Hoff, K. J., Lange, S., Lomsadze, A., Borodovsky, M. & Stanke, M. BRAKER1: unsupervised RNA-Seq-Based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32, 767–769 (2016).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 (2008).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Loveland, J. E., Gilbert, J. G., Griffiths, E. & Harrow, J. L. Community gene annotation in practice. Database (Oxford) 2012, bas009 (2012).
Pennisi, E. Ideas fly at gene-finding jamboree. Science 287, 2182–2184 (2000).
Archibald, A. L. et al. Pig genome sequence—analysis and publication strategy. BMC Genomics 11, 438 (2010).
Lee, E. et al. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 14, R93 (2013).
Giraldo-Calderon, G. I. et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43, D707–D713 (2015).
Dawson, H. D. et al. Structural and functional annotation of the porcine immunome. BMC Genomics 14, 332 (2013).
The UK 10K Consortium. The UK10K project identifies rare variants in health and disease. Nature 526, 82–90 (2015).
Guo, L., Gao, Z. & Qian, Q. Application of resequencing to rice genomics, functional genomics and evolutionary analysis. Rice (N.Y.) 7, 4 (2014).
Foote, A. D. et al. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat. Commun. 7, 11693 (2016).
Adams, D. J., Doran, A. G., Lilue, J. & Keane, T. M. The Mouse Genomes Project: a repository of inbred laboratory mouse strain genomes. Mamm. Genome 26, 403–412 (2015).
Baker, M. Structural variation: the genome's hidden architecture. Nat. Methods 9, 133–137 (2012).
Zarrei, M., MacDonald, J. R., Merico, D. & Scherer, S. W. A copy number variation map of the human genome. Nat. Rev. Genet. 16, 172–183 (2015).
Trowsdale, J. & Knight, J. C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genom. Hum. Genet. 14, 301–323 (2013).
Hirayasu, K. & Arase, H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J. Hum. Genet. 60, 703–708 (2015).
Iyer, M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015). In this study, thousands of human RNA-seq libraries are combined to generate almost 60,000 putative lncRNA genes.
Filichkin, S. A. et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58 (2010).
Mudge, J. M., Frankish, A. & Harrow, J. Functional transcriptomics in the post-ENCODE era. Genome Res. 23, 1961–1973 (2013).
Steijger, T. et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184 (2013).
Cho, H. et al. High-resolution transcriptome analysis with long-read RNA sequencing. PLoS ONE 9, e108095 (2014).
Tilgner, H. et al. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat. Biotechnol. 33, 736–742 (2015).
Mercer, T. R. et al. Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. Nat. Protoc. 9, 989–1009 (2014).
Jiang, L. et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 21, 1543–1551 (2011).
The FANTOM Consortium et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). The leading publication of the FANTOM5 project, providing detailed analysis of hundreds of human and mouse CAGE experiments.
Derti, A. et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 22, 1173–1183 (2012).
Boley, N. et al. Genome-guided transcript assembly by integrative analysis of RNA sequence data. Nat. Biotechnol. 32, 341–346 (2014).
Hezroni, H. et al. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 11, 1110–1122 (2015).
Sisu, C. et al. Comparative analysis of pseudogenes across three phyla. Proc. Natl Acad. Sci. USA 111, 13361–13366 (2014).
Frankish, A. & Harrow, J. GENCODE pseudogenes. Methods Mol. Biol. 1167, 129–155 (2014).
Carelli, F. N. et al. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 26, 301–314 (2016).
Zhang, Z. et al. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics 22, 1437–1439 (2006).
Pei, B. et al. The GENCODE pseudogene resource. Genome Biol. 13, R51 (2012).
Kelemen, O. et al. Function of alternative splicing. Gene 514, 1–30 (2013).
Yang, X. et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164, 805–817 (2016).
Pickrell, J. K., Pai, A. A., Gilad, Y. & Pritchard, J. K. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet. 6, e1001236 (2010).
Hao, Y. et al. Semi-supervised learning predicts approximately one third of the alternative splicing isoforms as functional proteins. Cell Rep. 12, 183–189 (2015).
Rodriguez, J. M. et al. APPRIS: annotation of principal and alternative splice isoforms. Nucleic Acids Res. 41, D110–D117 (2013).
Farrell, C. M. et al. Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res. 42, D865–D872 (2014).
Bassett, A. R. et al. Considerations when investigating lncRNA function in vivo. eLife 3, e03058 (2014).
Derrien, T., Guigo, R. & Johnson, R. The long non-coding RNAs: a new (P)layer in the “dark matter”. Front. Genet. 2, 107 (2011).
Hangauer, M. J., Vaughn, I. W. & McManus, M. T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 9, e1003569 (2013).
van Bakel, H., Nislow, C., Blencowe, B. J. & Hughes, T. R. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8, e1000371 (2010).
Peccarelli, M. & Kebaara, B. W. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot. Cell 13, 1126–1135 (2014).
Lareau, L. F. & Brenner, S. E. Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol. Biol. Evol. 32, 1072–1079 (2015).
Wong, J. J. et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154, 583–595 (2013).
Braunschweig, U. et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 24, 1774–1786 (2014). Demonstrates that intron retention affects three-quarters of mammalian genes, and suggests widespread involvement in gene regulation.
Capell, A., Fellerer, K. & Haass, C. Progranulin transcripts with short and long 5′ untranslated regions (UTRs) are differentially expressed via posttranscriptional and translational repression. J. Biol. Chem. 289, 25879–25889 (2014).
Barbosa, C., Peixeiro, I. & Romao, L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 9, e1003529 (2013).
Yeh, H. S. & Yong, J. Alternative polyadenylation of mRNAs: 3′-untranslated region matters in gene expression. Mol. Cells 39, 281–285 (2016).
Barrett, L. W., Fletcher, S. & Wilton, S. D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 69, 3613–3634 (2012).
Mudge, J. M. et al. The origins, evolution, and functional potential of alternative splicing in vertebrates. Mol. Biol. Evol. 28, 2949–2959 (2011).
Barash, Y. & Garcia, J. V. Predicting alternative splicing. Methods Mol. Biol. 1126, 411–423 (2014).
Nesvizhskii, A. I. Proteogenomics: concepts, applications and computational strategies. Nat. Methods 11, 1114–1125 (2014). An obvious starting point to explore strategies for the analysis of mass-spectrometry data in genomics.
Kim, M. S. et al. A draft map of the human proteome. Nature 509, 575–581 (2014).
Wilming, L. G. et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 36, D753–D760 (2008).
Ezkurdia, I., Vazquez, J., Valencia, A. & Tress, M. Analyzing the first drafts of the human proteome. J. Proteome Res. (2014).
Wright, J. C. et al. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat. Commun. 7, 11778 (2016).
Kim, T. K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Ingolia, N. T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 15, 205–213 (2014).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Jackson, R. & Standart, N. The awesome power of ribosome profiling. RNA 21, 652–654 (2015).
Ingolia, N. T. Ribosome footprint profiling of translation throughout the genome. Cell 165, 22–33 (2016). A primer on the use of RP from one of the key developers of the technique.
Raj, A. et al. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. eLife 5, e13328 (2016).
Mumtaz, M. A. & Couso, J. P. Ribosomal profiling adds new coding sequences to the proteome. Biochem. Soc. Trans. 43, 1271–1276 (2015).
Graur, D. et al. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol. 5, 578–590 (2013).
Xie, S. Q. et al. RPFdb: a database for genome wide information of translated mRNA generated from ribosome profiling. Nucleic Acids Res. 44, D254–D258 (2016).
Goff, L. A. & Rinn, J. L. Linking RNA biology to lncRNAs. Genome Res. 25, 1456–1465 (2015).
Palazzo, A. F. & Lee, E. S. Non-coding RNA: what is functional and what is junk? Front. Genet. 6, 2 (2015).
Kutter, C. et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 8, e1002841 (2012).
Ulitsky, I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17, 601–614 (2016).
Sleutels, F., Zwart, R. & Barlow, D. P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813 (2002).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Lai, F. & Shiekhattar, R. Enhancer RNAs: the new molecules of transcription. Curr. Opin. Genet. Dev. 25, 38–42 (2014).
Scruggs, B. S. et al. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol. Cell 58, 1101–1112 (2015).
Furio-Tari, P., Tarazona, S., Gabaldon, T. & Enright, A. J. & Conesa, A. spongeScan: A web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. (2016).
Novikova, I. V., Hennelly, S. P. & Sanbonmatsu, K. Y. Tackling structures of long noncoding RNAs. Int. J. Mol. Sci. 14, 23672–23684 (2013).
Konig, J., Zarnack, K., Luscombe, N. M. & Ule, J. Protein-RNA interactions: new genomic technologies and perspectives. Nat. Rev. Genet. 13, 77–83 (2011).
Quek, X. C. et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 43, D168–D173 (2015).
Volders, P. J. et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 43, 4363–4364 (2015).
Zhao, Y. et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 44, D203–D208 (2016).
RNAcentral Consortium. RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res. 43, D123–D129 (2015).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Belton, J. M. et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012).
Fullwood, M. J. & Ruan, Y. ChIP-based methods for the identification of long-range chromatin interactions. J. Cell Biochem. 107, 30–39 (2009).
Mifsud, B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015). This study uses Capture Hi-C to examine the long-range chromosome interactions of 22,000 human promoters.
Cairns, J. et al. CHiCAGO: robust detection of DNA looping interactions in capture Hi-C data. Genome Biol. 17, 127 (2016).
Dickel, D. E. et al. Function-based identification of mammalian enhancers using site-specific integration. Nat. Methods 11, 566–571 (2014).
Heinz, S., Romanoski, C. E., Benner, C. & Glass, C. K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015).
Shlyueva, D., Stampfel, G. & Stark, A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286 (2014).
Zerbino, D. R. et al. Ensembl regulation resources. Database (Oxford) 2016, 1–13 (2016).
de Wit, E. et al. CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015).
Ong, C. T. & Corces, V. G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014).
Gonzalez-Porta, M., Frankish, A., Rung, J., Harrow, J. & Brazma, A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 14, R70 (2013).
The GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Vogel, C. & Marcotte, E. M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 (2012).
Battle, A. et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science 347, 664–667 (2015).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Dalgleish, R. et al. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2, 24 (2010). This project provides insights into the relationship between gene annotation and the description of variation in the clinic.
Takahashi, H., Kato, S., Murata, M. & Carninci, P. CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol. Biol. 786, 181–200 (2012).
Batut, P., Dobin, A., Plessy, C., Carninci, P. & Gingeras, T. R. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 23, 169–180 (2013).
Fullwood, M. J. et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 (2009).
Johnson, D. S., Mortazavi, A., Myers, R. M. & Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 (2007).
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Carver, T., Harris, S. R., Berriman, M., Parkhill, J. & McQuillan, J. A. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469 (2012).
Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14, 178–192 (2013).
Benson, D. A. et al. GenBank. Nucleic Acids Res. 41, D36–D42 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Lin, M. F., Jungreis, I. & Kellis, M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–i282 (2011).
Nawrocki, E. P. et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137 (2015).
Kozomara, A. & Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014).
Yates, A. et al. Ensembl 2016. Nucleic Acids Res. 44, D710–D716 (2016).
Acknowledgements
The work performed by J.M.M. and J.H. on the GENCODE project is supported by the National Human Genome Research Institute of the National Institutes of Health (grant number U41 HG007234). The authors thank A. Frankish for informative discussions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
DATABASES
FURTHER INFORMATION
Glossary
- Gene
-
Redefined for the modern era by Gerstein et al. (Ref. 1) as “a union of genomic sequences encoding a coherent set of potentially overlapping functional products” (that is, RNAs or proteins).
- Genebuild
-
Used by GENCODE and Ensembl for a collection of transcript models generated by computational or manual annotation across an entire genome sequence. Protein-coding genes, long non-coding RNAs, small RNAs and pseudogenes may be included.
- Transcript
-
Any form of RNA molecule that is transcribed from the genome sequence.
- Functional annotation
-
The process of defining or predicting functional roles for transcript models during gene annotation.
- Alternative splicing
-
Process by which a gene makes distinct transcripts through the use of different splice sites or exon combinations; these are known as alternative transcripts or transcript variants.
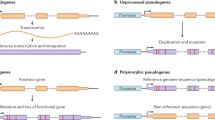
- Pseudogenes
-
'Broken' genes that are derived from protein-coding loci. Can be formed by retrotransposition ('processed'), duplication ('unprocessed') or inactivation ('unitary', which may be polymorphic). All forms may be transcribed.
- Long non-coding RNAs
-
(lncRNAs). Genes that do not contain protein-coding transcripts and that are not pseudogenes or small RNAs; a 200 bp size cut-off is typically applied to distinguish them from small RNAs.
- Small RNA
-
A member of one of several known families of small RNA molecules. Includes the classic tRNA and rRNA families alongside more recent discoveries such as PIWI-interacting RNAs (piRNAs), microRNAs (miRNAs) and small nucleolar RNAs (snoRNAs).
- Coding sequences
-
(CDSs). The regions of a transcript that are translated, that is, contain the information that encodes a protein sequence.
- Manual annotation
-
When a person constructs a transcript model de novo after appraising the available evidence (typically using software tools), or examines and potentially validates ('curates') a model that has been created computationally.
- Computational annotation
-
The process of generating genebuilds through entirely in silico processes, that is, by the use of computational algorithms.
- Transcription start site
-
(TSS). The base pair on the genome where transcription begins.
- Polyadenylation tail
-
A sequence of adenosine monophosphates attached to the 3′ end of an RNA as transcription terminates, beginning at the polyA site.
- Translation initiation site
-
(TIS). The codon that is translated to give the first amino acid of a peptide; almost always ATG; also known as a START codon.
- STOP codon
-
The final codon of a protein translation; almost always TAG, TAA or TGA; also known as a translation termination site or codon.
- Isoforms
-
Protein molecules that differ in their amino acid composition from other translations made from the same gene, for example, owing to alternative splicing.
- Intron retention
-
Occurs when a transcript does not splice out one or more introns, that is, this sequence is left incorporated into the mature RNA.
- Nonsense-mediated decay
-
(NMD). Cellular 'surveillance' mechanism that targets transcripts for destruction. Imprecisely understood, although transcripts featuring termination codons more than 50 bp upstream of splice junctions are thought likely to be substrates.
- Poison exon
-
An exon that prevents correct coding sequence translation when incorporated into the transcript of a protein-coding gene, either by causing a frameshift or through the introduction of a premature termination codon.
- Untranslated region
-
(UTR). Non-coding sequence on coding sequence transcripts found between the transcription start site and the translation initiation site (5′ UTR), and the STOP codon and polyA site (3′ UTR).
- Enhancer
-
Sequence that regulates a promoter from a distal site on the chromosome, probably brought into close proximity through DNA looping.
- Promoters
-
Regions immediately upstream of the transcription start site where the RNA polymerase complex attaches in order to initiate transcription.
Rights and permissions
About this article
Cite this article
Mudge, J., Harrow, J. The state of play in higher eukaryote gene annotation. Nat Rev Genet 17, 758–772 (2016). https://doi.org/10.1038/nrg.2016.119
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2016.119
This article is cited by
-
Transformation of alignment files improves performance of variant callers for long-read RNA sequencing data
Genome Biology (2023)
-
Enhanced protein isoform characterization through long-read proteogenomics
Genome Biology (2022)
-
Bioinformatic approach for the discovery of cis-eQTL signals during fruit ripening of a woody species as grape (Vitis vinifera L.)
Scientific Reports (2022)
-
Dominant transcript expression profiles of human protein-coding genes interrogated with GTEx dataset
Scientific Reports (2022)
-
Understanding the causes of errors in eukaryotic protein-coding gene prediction: a case study of primate proteomes
BMC Bioinformatics (2020)