Abstract
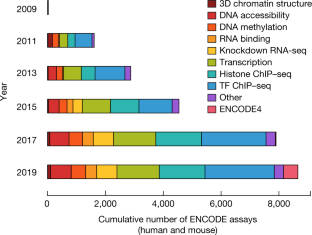
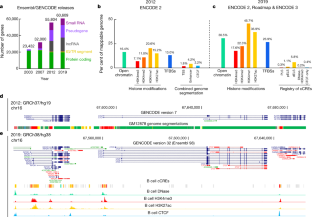
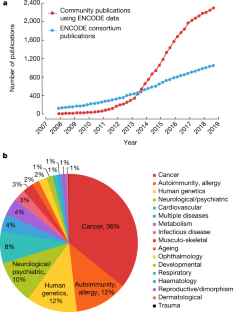
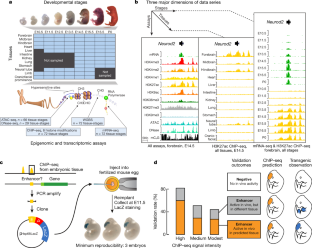
The Encylopedia of DNA Elements (ENCODE) Project launched in 2003 with the long-term goal of developing a comprehensive map of functional elements in the human genome. These included genes, biochemical regions associated with gene regulation (for example, transcription factor binding sites, open chromatin, and histone marks) and transcript isoforms. The marks serve as sites for candidate cis-regulatory elements (cCREs) that may serve functional roles in regulating gene expression1. The project has been extended to model organisms, particularly the mouse. In the third phase of ENCODE, nearly a million and more than 300,000 cCRE annotations have been generated for human and mouse, respectively, and these have provided a valuable resource for the scientific community.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Change history
26 April 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41586-021-04213-8
References
Kellis, M. et al. Defining functional DNA elements in the human genome. Proc. Natl Acad. Sci. USA 111, 6131–6138 (2014).
ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 (2004).
Lindblad-Toh, K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011).
Waterston, R. H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9, e1001046 (2011).
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007). The results of the pilot phase of ENCODE included extensive functional assays across a selected one per cent of the human genome with experiments conducted on a variety of cell lines and largely with array-based technology.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). The results of the second phase of ENCODE were based mostly on a large number of genome-wide assays that leveraged high-throughput sequencing technologies and were done across two ‘tier one’ cell lines with large-scale assays across several hundred cell and tissue types.
The ENCODE Project Consortium et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature https://doi.org/10.1038/s41586-020-2493-4 (2020).
Partridge, E. C. et al. Occupancy maps of 208 chromatin-associated proteins in one human cell type. Nature https://doi.org/10.1038/s41586-020-2023-4 (2020).
Meuleman, W. Index and biological spectrum of human DNase I hypersensitive sites. Nature https://doi.org/10.1038/s41586-020-XXXX-X (2020).
Vierstra, J. et al. Global reference mapping of human transcription factor footprints. Nature https://doi.org/10.1038/s41586-020-2528-x (2020).
Breschi, A. et al. A limited set of transcriptional programs define major cell types. Preprint at https://doi.org/10.1101/857169 (2020).
Grubert, F. et al. Landscape of cohesin-mediated chromatin loops in the human genome. Nature https://doi.org/10.1038/s41586-020-2151-x (2020).
Van Nostrand, E. L. et al. A large-scale binding and functional map of human RNA binding proteins. Nature https://doi.org/10.1038/s41586-020-2077-3 (2020).
Wang, Z., Gerstein, M. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 (2008).
Johnson, D. S., Mortazavi, A., Myers, R. M. & Wold, B. Genome-wide mapping of in vivo protein–DNA interactions. Science 316, 1497–1502 (2007).
Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4, 651–657 (2007).
Iyer, V. R. et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409, 533–538 (2001).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Landt, S. G. et al. ChIP–seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). A consortium-wide effort to standardize performance, quality control and outputs of ChIP–seq experiments, including validation of antibodies, to facilitate experimental reproducibllity and data utility.
Sundararaman, B. et al. Resources for the comprehensive discovery of functional RNA elements. Mol. Cell 61, 903–913 (2016).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Schaub, M. A., Boyle, A. P., Kundaje, A., Batzoglou, S. & Snyder, M. Linking disease associations with regulatory information in the human genome. Genome Res. 22, 1748–1759 (2012).
Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014). Results of a large-scale effort of the mouse ENCODE consortium, presenting regulatory and transcript maps of the mouse.
Gerstein, M. B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787 (2010).
The modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Kudron, M. M. et al. The ModERN Resource: genome-wide binding profiles for hundreds of Drosophila and Caenorhabditis elegans transcription factors. Genetics 208, 937–949 (2018).
Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature https://doi.org/10.1038/s41586-020-2093-3 (2020).
He, P. A. The changing mouse embryo transcriptome at whole tissue and single-cell resolution. Nature https://doi.org/10.1038/s41586-020-XXXX-X (2020).
He, Y. et al. Spatiotemporal DNA methylome dynamics of the developing mouse fetus. Nature https://doi.org/10.1038/s41586-020-2119-x (2020).
Cheng, Y. et al. Principles of regulatory information conservation between mouse and human. Nature 515, 371–375 (2014).
Stefflova, K. et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell 154, 530–540 (2013).
Keilwagen, J., Posch, S. & Grau, J. Accurate prediction of cell type-specific transcription factor binding. Genome Biol. 20, 9 (2019).
Tang, F., Lao, K. & Surani, M. A. Development and applications of single-cell transcriptome analysis. Nat. Methods 8 (Suppl), S6–S11 (2011).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Hu, B. C.; HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Regev, A. et al. The human cell atlas. eLife 6, e27041 (2017).
Rhoads, A. & Au, K. F. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13, 278–289 (2015).
Arnold, C. D. et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (2013).
Klein, J. C., Chen, W., Gasperini, M. & Shendure, J. Identifying novel enhancer elements with CRISPR-based screens. ACS Chem. Biol. 13, 326–332 (2018).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Paudyal, A. et al. The novel mouse mutant, chuzhoi, has disruption of Ptk7 protein and exhibits defects in neural tube, heart and lung development and abnormal planar cell polarity in the ear. BMC Dev. Biol. 10, 87 (2010).
Acknowledgements
We thank S. Moore, E. Cahill, M. Kellis and J. Li for their assistance, and B. Wold for helpful comments. This work was supported by grants from the NIH: U01HG007019, U01HG007033, U01HG007036, U01HG007037, U41HG006992, U41HG006993, U41HG006994, U41HG006995, U41HG006996, U41HG006997, U41HG006998, U41HG006999, U41HG007000, U41HG007001, U41HG007002, U41HG007003, U41HG007234, U54HG006991, U54HG006997, U54HG006998, U54HG007004, U54HG007005, U54HG007010 and UM1HG009442.
Author information
Authors and Affiliations
Consortia
Contributions
The role of the NHGRI Project Management Group in the preparation of this paper was limited to coordination and scientific management of the ENCODE consortium. All other authors contributed to the concepts, writing and/or revisions of this manuscript.
Corresponding author
Ethics declarations
Competing interests
B.E.B. declares outside interests in Fulcrum Therapeutics, 1CellBio, HiFiBio, Arsenal Biosciences, Cell Signaling Technologies, BioMillenia, and Nohla Therapeutics. P.F. is a member of the Scientific Advisory Boards of Fabric Genomics, Inc. and Eagle Genomics, Ltd. M.P.S. is cofounder and scientific advisory board member of Personalis, SensOmics, Mirvie, Qbio, January, Filtricine, and Genome Heart. He serves on the scientific advisory board of these companies and Genapsys and Jupiter. Z.W. is a cofounder of Rgenta Therapeutics and she serves on its scientific advisory board. R.M.M. is an advisor to DNAnexus and Decheng Capital, and has outside interests in IMIDomics, Accuragen and ReadCoor, Inc. The authors declare no other competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 ENCODE timeline.
Pilot phase: September 2003–September 2007; ENCODE 2: September 2007–September 2012; ENCODE 3: September 2012–January 2017; ENCODE 4: February 2017–present; modENCODE: April 2007–April 2012; mouse ENCODE: 2009–2012.
Supplementary information
Supplementary Information
This file contains the full author list for The ENCODE Project Consortium, and Supplementary Note 1 (Useful URLs).
Rights and permissions
About this article
Cite this article
The ENCODE Project Consortium., Snyder, M.P., Gingeras, T.R. et al. Perspectives on ENCODE. Nature 583, 693–698 (2020). https://doi.org/10.1038/s41586-020-2449-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2449-8
This article is cited by
-
How much do we know about the function of mammalian genes?
BMC Biology (2023)
-
Bioinformatics for wet-lab scientists: practical application in sequencing analysis
BMC Genomics (2023)
-
How rare mutations contribute to complex traits
Nature (2023)
-
Interpreting non-coding disease-associated human variants using single-cell epigenomics
Nature Reviews Genetics (2023)
-
Lack of GPR180 ameliorates hepatic lipid depot via downregulation of mTORC1 signaling
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.



