Abstract
Idiopathic pulmonary fibrosis (IPF) is a severe and progressive chronic fibrosing interstitial lung disease with causes that have remained unclear to date. Development of effective treatments will require elucidation of the detailed pathogenetic mechanisms of IPF at both the molecular and cellular levels. With a biomedical corpus that includes IPF-related entities and events, text-mining systems can efficiently extract such mechanism-related information from huge amounts of literature on the disease. A novel corpus consisting of 150 abstracts with 9297 entities intended for training a text-mining system was constructed to clarify IPF-related pathogenetic mechanisms. For this corpus, entity information was annotated, as were relation and event information. To construct IPF-related networks, we also conducted entity normalization with IDs assigned to entities. Thereby, we extracted the same entities, which are expressed differently. Moreover, IPF-related events have been defined in this corpus, in contrast to existing corpora. This corpus will be useful to extract IPF-related information from scientific texts. Because many entities and events are related to lung diseases, this freely available corpus can also be used to extract information related to other lung diseases such as lung cancer and interstitial pneumonia caused by COVID-19.
Similar content being viewed by others
Introduction
Idiopathic pulmonary fibrosis (IPF), a severe chronic fibrosing interstitial lung disease of unclear etiology, characteristically leads to progressive and irreversible decline of lung function1. Idiopathic pulmonary fibrosis (IPF) acute exacerbation is a serious condition with acute respiratory failure, and representative studies have shown a 30-day survival rate of 44.6% and a 90-day survival rate of 24.6% after hospitalization for developing IPF acute exacerbation2. In addition, there are reports of significant fibrosis progression even after recovery, making prevention of acute exacerbations an important aspect of IPF management3. Although medications such as pirfenidone and nintedanib have been used to slow the progression of IPF, no medical treatment can cure IPF completely1,2,3,4,5. Pirfenidone is an antifibrotic and anti-inflammatory drug4,6. Nintedanib, an intracellular kinase inhibitor, targets multiple tyrosine kinases such as vascular endothelial growth factor (VEGF) receptor, fibroblast growth factor (FGF) receptor, and platelet-derived growth factor (PDGF) receptor5. Developing more efficient medications that can fundamentally treat the disease will necessitate elucidation of the detailed pathogenetic mechanisms of IPF at both molecular and cellular levels.
More than a hundred thousand reports of the literature on IPF have been registered in the PubMed database7: the most widely used online bibliographic database serving the biological sciences8. However, the availability of trained annotators with IPF-related knowledge is limited. Extracting adequate IPF-related information, and that of related phenomena (or ‘events’) and clinical processes, and effects of clinical treatments, from such huge amounts of information can be expected to be time-consuming. Consequently, efficient text-mining methods must be used to extract adequate information from the copious literature.
Text-mining systems have been developed for biomedical research, with information extraction algorithms and corpora corresponding particularly to systems biology, for which pathways and networks are often constructed9. Particularly, systems such as NERsuite10 and EventMine11,12, which employ traditional feature-based machine learning methods, have been used to extract biomedical entities and events (or phenomena) from such corpora. Recently, a neural event extraction model that employs deep learning has been proposed: DeepEventMine13. It shows higher performance in extracting events from such corpora. Biomedical corpora that include biomedical events have been constructed: GENIA14,15, Gene Regulation Event Corpus (GREC)16, and Cancer Genetics corpus17,18,19. In these corpora, genes and gene products (GGPs) as named entities have been annotated, along with events involving GGPs, such as gene expression and binding. Some entities and events related to IPF are annotated in the existing corpora. Nevertheless, none of these corpora are specifically associated with IPF. Information in the existing corpora is insufficient to construct IPF-related networks. Entity-linking, for which IDs must be assigned to entities, is necessary to normalize the same entities expressed differently. However, those existing corpora do not always have entity normalization. Furthermore, disease-related events have not been defined for the existing corpora, leading to difficulty in extracting disease-related events.
This work particularly examines the annotation of IPF-related entities, events, and relations to facilitate the automatic extraction of IPF-related information from scientific texts. After defining a new annotation schema for IPF-related abstracts, including the definitions of entities, events, and relations, we apply the schema and use the brat rapid annotation tool to annotate a corpus of 150 abstracts selected by experts on IPF20,21. Using the information in the existing corpora during the corpus development would be helpful, but the general methodologies to reuse existing corpora for the new annotation target have not been established yet. To avoid any difficulty in the annotation process, we annotate IPF-related entities, relations, and entities without relying on the existing corpora except for the automatic annotation toolkit, details of which will be described herein.
Methods and materials
For this work, the types of entities, events, and relations, and the UMLS semantic types, which will be described below, are double-quoted. Those annotated words and phrases in text data are single-quoted, whereas event arguments, also described below, are single-quoted in italic.
Definition of IPF-related entities
We defined essential entities involved in IPF-related phenomena and clinical processes (Table 1). Most biological entities were defined based on the GENIA meta-knowledge corpus22,23 and the PHAEDRA corpus24,25.
First, the “Disorder” entity was defined to extract information related to disease, injury, and symptoms. These entities were categorized together because it is difficult and time-consuming for annotators to distinguish diseases and injuries from symptoms. “Measurement” entity was also defined for the named entity of quantification for lung diseases. For instance, ‘Forced vital capacity (FVC)’, which is measured by spirometry, can be included in this category. “Subject” was defined for patients, subjects for clinical trials, and animals used for experimentation, indicating the whole-body level.
As for the sub-whole-body level, “Anatomical_entity”, “Cell”, and “Cell_component” were defined (Table 1). Organs and tissues are categorized in “Anatomical_entity”. Entities such as ‘serum’ and ‘Bronchoalveolar Lavage Fluid’, the UMLS semantic types of which fall into “body substance”26,27,28, were also included in “Anatomical_entity” for this corpus. Cell types and cell lines are included in “Cell”. Herein, “Cell_component” is defined for cellular components such as cytoplasm, transmembranes, and organelles.
Molecular entities consist of “Pharmacological_substance”, “GGPs”, “Organic_compound_other”, and “Inorganic_compound” (Table 1). “Pharmacological_substance” is defined for medicines. “GGPs” is defined for genes or gene products. These entities were categorized together as “GGPs” because it is difficult and time-consuming for annotators to discern genes and gene products such as gene transcripts, mRNA, and proteins, in text data. Earlier, such a gene-tag annotation as “GGPs” had been proposed for other biological corpora29,30. “Organic_compound_other” is defined for organic compounds, excluding medicines, genes, and gene products, whereas “Inorganic_compound” denotes inorganic substances such as metal ions.
“Entity_Property” and “Genetic_info” are defined for entities that cannot be included among the entities described above (Table 1). In “Entity_Property”, other technical terms, which include the degree of disease progression/stage, cell cycle stages, and attributes, such as immunophenotyping, for cells or genes, can be categorized. Mutation information for genes is categorized as “Genetic_info”.
In addition to the entities described above, we defined cue entities “Negation_cue” and “Speculation_cue” to indicate negation or confirmation and speculation degree for events, as described below. Negation words such as ‘no’, ‘not’, and ‘none’ can be a “Negation_cue”, whereas verbs such as ‘suggest’, ‘show’, and ‘indicate’, and auxiliary verbs such as ‘may’ and ‘might’ can be included as a “Speculation_cue”. The objective of “Negation_cue” is the same as that of the Negative Polarity, which can indicate negated events, in the GENIA meta-knowledge corpus22,23. In addition to these two cues, “Method_cue” was defined to indicate the type of experimental study and clinical examination. “Method_cue” might also suggest confirmation and degree of speculation about an event. Named entities such as ‘CT scans’ and ‘RT-PCR’ can be categorized in this cue. These cues are usually combined with event trigger words, as described below.
Definition of events for IPF
We defined artificial and biological events as presented in Table 2. Although only one artificial event was defined, biological events of several types were defined (Table 2 and Fig. 1). Most biological events were defined similarly to those in the GENIA meta-knowledge corpus22,23. Actually, biological events can be categorized into several events such as “Regulation”, “Correlation”, “Cellular_process”, and “Molecular_function”. Main components of these events are defined as the ‘triggers’ (or ‘trigger words’). ‘Triggers’ are expressed in various ways: verbal ones (e.g. ‘inhibit’), nominalizations of verbs (e.g. ‘inhibition’), and functional roles (noun) (e.g. ‘inhibitor’), in the case of ‘inhibition’ for “Negative_regulation” events. Each ‘trigger’ can be combined with major arguments, such as ‘Theme’, ‘Cause’, and ‘Participant’ along with auxiliary arguments such as ‘atLoc’ and ‘disorder’ (Table 2). In contrast to the other arguments, ‘disorder’ is a novel argument defined for our corpus. With the ‘disorder’ argument, ‘disorder’-related events (Fig. 1e–j) can be annotated separately from events that are not related to ‘disorder’ (Fig. 1a–d).
Annotation examples shown in format of brat rapid annotation tool. “Artificial_process” event (a), “Biological_process” event (b), “Negative_regulation” event (c), “Correlation” event with two “Negative_regulation” events and “Positive_regulation” event (d), ‘disorder’-related “Gene_expression” events (e), ‘disorder’-related “Positive_regulation” events with “Cellular_process” events (f), ‘disorder’-related “Positive_regulation” event with “Gene_expression” event (g), ‘disorder’-related “Positive_regulation” events (h), ‘disorder’-related “Positive_regulation” events with “Biological_process” event (i) and ‘disorder’-related “Negative_regulation” event with “Artificial_process” event, “Cellular_process” event and “Positive_regulation” event (j).
“Regulation” events, which suggest causality (cause and effect), are classifiable into two types: “Positive_regulation”, which describes ‘activation/up-regulation’ events, and “Negative_regulation”, which describes ‘inactivation/inhibition/down-regulation’ events. However, if it is not clear whether those trigger words are positive or negative, the “Regulation” event will be selected. Regarding arguments for “Regulation” events, what induces these “Regulation” events can be annotated as a ‘Cause’ argument, whereas the effect or target can be annotated as a ‘Theme’ argument, as presented in Fig. 1c,f,h,i,j.
In contrast to the “Regulation” events, the “Correlation” event was also defined because causalities are unclear in many cases. When several events and entities are correlated, these “Correlation” events will be adopted. Alternatively, when several events occur simultaneously, such events can be connected with this “Correlation” event. In contrast to the “Regulation” event, more than two events or entities as ‘Theme’ arguments can be associated with the “Correlation” event (Fig. 1d). In the case portrayed in Fig. 1d, one “Positive_regulation” event and two “Negative_regulation” events are associated with the “Correlation” event. With these events of two types, “Regulation” and “Correlation”, the annotated entities and events can be connected to develop a network of information. The earlier reported corpus for biological events, the GENIA corpus, also includes events of both types: “Regulation” and “Correlation”14,15,23. However, only the “Regulation” event is defined in the Cancer Genetics corpus17,18,19.
In addition to the “Regulation” and “Correlation” events, other biological events are categorized in “Localization”, “Cellular_process”, and “Molecular_function”. Among “Molecular_function” events, more specific molecular events are further classified into “Pathway”, “Conversion”, “Gene_expression”, “Binding”, and “Dissociation”. The “Localization” event describes localization and movement of entities such as “Cell” and molecular entities including “GGPs”. The “Pathway” describes signaling transduction or metabolic pathways, where molecular entities such as “GGPs” are involved as ‘Participant’. The “Conversion” event describes specific reactions that involve a change in covalent bonds. ‘Phosphorylation’ is an example of a “Conversion” event. “Gene_expression” describes either transcription or translation, for which only the “GGPs” entity can be annotated as ‘Theme’. Although “Binding” and “Dissociation” were also defined for molecular interaction and dissociation, it turned out that there are few cases for “Binding” and none for “Dissociation” (Table 2).
Event modifications such as ‘Negated’ events and ‘Speculated’ events were also defined. The events which can be connected with “Negation_cue” are defined as ‘Negated’ events, whereas those events which can be connected with “Speculation_cue” are defined as ‘Speculated’ events. These event modifications had already been defined in other corpora such as those for Cancer Genetics and Pathway Curation19. Moreover, the ‘Negated’ events are the same as those ‘negated bio-events’ defined by Nawaz et al.31. They are also similar to ‘Negative polarity’ defined by Thompson et al.23.
Normalization of entities/event triggers
The same named entities, which are often expressed differently, should be normalized to extract information properly from text data. For this work, normalization processing was performed by assigning the same ID to the same entities, which are expressed differently. Regarding such IDs, those for the Unified Medical Language System (UMLS) database (version 2018AB)26,27 were adopted for automatic annotation by MetaMap Lite32,33, which will be described below, and for the database installed in the brat annotation system20,21, with which the annotated IDs for UMLS were corrected manually after automatic annotation. The NCI Metathesaurus34, based on the UMLS database, was also used for manual annotation because the annotators had to search manually for the most appropriate terms when exact terms were not detected in the UMLS database installed in the brat system. Furthermore, event triggers were normalized along with entities.
Definition of relations for IPF
We also defined some relations to represent static relations between entities and events. Such relations include “part_of”, “member_of”, “Subject_Disorder”, and “Disorder_association” (Table 3).
The “part_of” relation can indicate relations of a partial entity with a whole entity, which is constituted by the partial entity. For example, this relation can indicate the relations between “Cell” and “Anatomical_entity”, such as tissues and organs. It is extremely useful to extract such relations from text data. The “member_of” relations can indicate a relation of a member with a group to which the member belongs. For example, this relation can indicate relations between a protein and its protein families, and between a patient and a patient group.
“Subject_Disorder” was defined to relate “Subject” and “Disorder”, following the relation defined in the PHAEDRA corpus24,25. “Disorder_association” was defined to indicate complications of diseases. Complications by two “Disorders” can be annotated by connecting the corresponding “Disorder” entities with “Disorder_association”.
Annotation process
Selection of abstracts for annotation
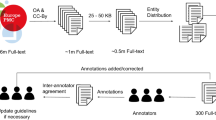
We constructed the corpus composed of 150 abstracts of research articles on IPF-related basic research involving molecular biology. A lung disease expert manually selected the 150 abstracts: first, we narrowed down the number of IPF-related articles to about 6500 from about 100,000 articles in major journals registered in PubMed from 2013 to 2018, and selected 500 articles included in the categories of preclinical, with drugs such as pirfenidone, nintedanib, dexamethasone, tacrolimus, fluorofenidone, sirolimus, leflunomide, azithromycin, β-lapachone, sunitinib, carnosine, and tamoxifen, and without drugs. After preliminary curation to ensure that a sufficiently diverse group of molecules was included, we narrowed the list further to prioritize those with sufficiently detailed abstracts and rich descriptions: those which included descriptions of molecules and pathways associated with various respiratory diseases such as IPF and lung cancer, such as ‘TGF-β’, ‘Surfactant protein’, ‘signaling pathway’, ‘migration’, ‘macrophage’, ‘MMP’, ‘CTGF’, and ‘mucin’.
Automatic annotation, which is described in the next section, was conducted for the abstracts of the top 300 articles that were prioritized manually as described above. From the 300 automatically annotated abstracts, 120 abstracts were selected randomly for manual annotation. Moreover, from the remainder of the abstracts for inter-annotator agreement (IAA), 30 abstracts that contained numerous GGPs were selected to increase the cases of molecular events.
Automatic annotation by MetaMap Lite and UMLS semantic types
The MetaMap Lite 3.6.2rc3 and UMLS 2018AB datasets were applied to perform automatic annotation for the selected abstract dataset26,27,32,33. MetaMap Lite is a Java implementation of the basic functions of MetaMap35,36, which is a named entity recognition (NER) tool able to identify Unified Medical Language System (UMLS) Meta-thesaurus concepts28 in biomedical texts. Actually, MetaMap Lite can provide the longest concept-matched words and phrases with the UMLS concept unique identifier (CUI), designated herein as ‘UMLS ID’, as well as an “MMLite” tag. Each UMLS CUI has at least one semantic type such as “dsyn; Disease and Syndrome” and “gngm; Gene or Genome”.
The tags for the entity types, which are defined and described above, were assigned based on the semantic types. The “MMLite” tags were replaced with those tags for the entity types. For example, the “Disorder” entity tag will be assigned to the concept-matched words and phrases for the semantic type, “dsyn; Disease and Syndrome”, whereas the “GGPs” entity tag will be assigned for “gngm; Gene or Genome”. However, when a CUI (UMLS ID) is associated with multiple semantic types, selecting one automatically can engender the assignment of an unsuitable tag for the context. In such cases, annotators must consider and correct the predicted annotated entities manually.
Manual annotation: guideline construction and annotators
The manual annotation process used for this work is presented in Fig. 2. To develop a consistent corpus, the annotation leader, a protein researcher with experience in text-annotation, constructed the annotation guideline for all annotators using the Annodoc documentation support system37,38. The Annodoc system is useful for constructing guidelines for text-annotation because it can readily include annotation examples in the brat format. The annotation scheme used for brat tool configuration was designed by the guideline author.
Manual annotation process for the corpus. The annotation leader constructed the annotation guideline. Based on the guideline, the annotation leader also designed the annotation scheme and the configuration for the brat tool. The annotators did text-annotation manually. The guideline was revised occasionally based on the annotation data and scheme.
Manual annotation was conducted by an annotator (annotator 1) using the brat rapid annotation tool20,21. Annotator 1 has experience in the translation of biomedical documents. To ensure inter-annotator agreement (IAA), another annotator (annotator 2) performed manual annotation for 30 selected abstracts. Annotator 2 is a protein researcher specializing in signaling pathways, with experience in text-annotation for signaling pathways. The IAA dataset produced by the two annotators is available39.
Moreover, annotation meetings were held occasionally among the guideline author, the annotators, and the IPF expert to discuss difficult annotations. The guideline was revised based on those discussions. Also, the annotation was corrected. The guideline is available40.
Evaluation
Evaluation of this corpus was based on the standard metrics of precision, recall, and F1-score. We applied the automatic entity detection and event extraction methods to the corpus and evaluated its performance. We used an event extraction system, DeepEventMine13, and a neural named entity recognition and linking system, BERT-based Exhaustive Neural Named Entity Recognition and Disambiguation (BENNERD)41.
DeepEventMine, a neural end-to-end event extraction model, extracts events from raw sentences. It performs trigger and entity recognition, relation classification, and event detection in an end-to-end manner. As another neural model, BENNERD consists of a span-based exhaustive named entity recognition model and an entity-linking model. The entity-linking model performs candidate generation that identifies a list of candidate entities in UMLS for a given mention and candidate ranking that ranks the entity candidate list to choose the best entity for the mention.
After separately evaluating event triggers and entities, entity-linking, relations and events, we used BENNERD to train individual entity recognition and linking models for each trigger and entity type. For relations, we used the trigger and entity recognition and relation extraction modules in DeepEventMine. We performed ten-fold cross-validation and measured the F1-scores with exact boundary matching for triggers, entities, and relations. For event extraction, we applied DeepEventMine, and followed the evaluation protocol adopted by BioNLP Shared Task 200942 to evaluate our event prediction. In practice, we adopted the evaluation script introduced into the Cancer Genetics 201318. Then we calculated the F1-scores of detected event structures using the primary matching criteria in the task.
Measurement of inter-annotator agreement (IAA) was performed using the same evaluation criteria as those used for the automatic evaluation explained above (i.e., F1 scores). We calculated the F1 scores by treating the annotations of one annotator as a gold standard and those of the other annotator as a system prediction. We switched the roles of the two annotators and averaged the F1 scores to obtain the final IAA scores. To evaluate IAA of entity-linking annotations, we only considered entities and triggers shared by the two annotators and evaluated linking annotations. Similarly, for relations, we evaluated IAA of relation types among triggers and entities shared by the two annotators to evaluate IAA based on relations alone. Regarding events, we considered entities shared by two annotators as gold entities and ignored the remaining entities and evaluated IAA.
Results and discussion
Tendencies in corpus contents
The corpus developed for this work was analyzed. Despite the small number of documents, only 150 abstracts, the total number of entities annotated in the corpus was 8524 (without including the three cues in Table 1), which is comparable to earlier-developed corpora such as the multi-level event extraction (MLEE) corpus, with 8291 entities43. Table 1 shows that “GGPs”, “Disorder”, “Subject”, “Anatomical_entity”, and “Cell” were observed frequently among all entities. The frequently observed UMLS IDs and their respective references were analyzed for the entities (Table 4).
For “Disorder”, it is natural that the UMLS ID indicating ‘Idiopathic Pulmonary Fibrosis’ was the most frequently observed along with those for other lung diseases (Table 4a). In addition to these IDs for lung diseases, the UMLS ID for ‘Sarcoidosis’ was observed frequently (Table 4a). In the lungs of ‘Sarcoidosis’, the disease follows the pathology of interstitial pneumonia, and if the inflammation persists, pulmonary fibrosis may occur, limiting activity and interfering with daily life due to cough and shortness of breath. In this point of view, ‘Sarcoidosis’ appears with certain frequency. There are regional and racial differences in the incidence and severity of the disease, for example, in Europe it is more common in Northern Europe than Southern Europe44, and in the USA, black races are several times more susceptible and severely affected than Caucasians45. In Japan, by gender, twice as many women as men are detected and by age, the disease is bimodal in both men and women, in their 20 s and after their 50 s46. In this study, because of the focus on the respiratory tract, terms related to pulmonary fibrosis in sarcoidosis were extracted, but not terms related to the epidemiological differences described above.
Thirty-five of “Disorder” entities, which correspond to 'combined pulmonary fibrosis and emphysema', 'familial pulmonary fibrosis', 'unilateral ureteral obstruction renal fibrosis', 'non-infectious disease' and 'canine idiopathic pulmonary fibrosis', could not be assigned UMLS IDs to (Table 4a). In this corpus, 'canine idiopathic pulmonary fibrosis' was distinguished from human ‘IPF’ without being assigned the same ID.
Regarding “Measurement”, the UMLS IDs for measurements of pulmonary function and neutrophil were observed frequently (Table 4b). The UMLS IDs for ‘Patients’ and ‘Control group’ were observed most frequently for “Subject” (Table 4c).
For “Anatomical_entity”, the UMLS IDs for ‘Lung’ and ‘Serum’ were the most frequently observed, indicating that these two IDs appear once in each abstract (Table 4d). ‘Bronchoalveolar Lavage Fluid’, for which the UMLS semantic type falls into “body substance”, was also observed frequently for “Anatomical_entity” (Table 4d). Regarding “Cell”, the UMLS IDs for fibroblasts, epithelial cells, leukocytes such as neutrophils, lymphocytes, and macrophages were observed frequently (Table 4e). Although the total number of “Pharmacological_substance” is rather low (Table 1), ‘Bleomycin’, which is used to induce and model pulmonary fibrosis, medicine for IPF, ‘FG 3019’, an expectorant, ‘Acetylcysteine’, and mTOR inhibitor, such as ‘Sirolimus’, were often observed (Table 4f). Regarding “GGPs”, the UMLS IDs for cytokines and growth factors were observed frequently (Table 4g). Among the cytokines, ‘Interleukin-8’, which induces chemotaxis in target cells, was the most frequently observed (Table 4g). There were 79 “GGPs” entities with no UMLS IDs, because these entities indicate fragments, siRNA, or antibodies for some specific proteins, or ‘factors’ and ‘mediators’ that are not any specific “GGPs”, which do not have any UMLS IDs (Table 4g).
The events annotated in the corpus were 4899 (Table 2), which is a comparable number to those of some earlier developed corpora such as the MLEE corpus (6677 events)43, the epigenetic and post-translational modification (EPI) corpus (3714 events), and the infectious disease (ID) corpus (4150 events), which were developed by BioNLP Shared Task 201147.
As shown in Table 2, “Positive_regulation” and “Biological_process” were observed most frequently among all the defined events, although the occurrences of “Binding” and “Dissociation” were very few. The frequently observed UMLS IDs and their respective references were also analyzed for event trigger words (Table 5).
Regarding the trigger words for “Artificial_process”, the UMLS IDs for clinical actions, such as ‘Therapeutic procedure’, ‘Administer’ and ‘Diagnosis’, were most-frequently observed (Table 5a). Regarding “Biological_process”, high-order phenomena, or high-order events, such as pathogenesis, exacerbation and progression of disease, ‘Fibrosis’, and ‘Inflammation’, were observed frequently (Table 5b). ‘Exacerbation acute’ was detected as “Biological_process” event 72 times (Table 5b), of which 49 ‘Themes’ were IPF, for which ‘surgical lung biopsy’ of “Artificial_process” was detected as ‘Cause’ only once. Although the event trigger, ‘progressive respiratory failure’, was not identified in this corpus, ‘Disease Progression’ was detected 35 times, instead of such an event (Table 5b). For the ‘Disease Progression’, several “Disorder” types, and a few “Biological_process” were detected as ‘Theme’, among which IPF appeared 9 times. Regarding trigger words for “Localization” event, the UMLS IDs for secretion, accumulation, and cell migration were observed frequently (Table 5c). Regarding “Cellular_process”, the UMLS ID for ‘Cell Proliferation’ and ‘epithelial to mesenchymal transition (EMT)’ were observed most frequently (Table 5d). The EMT is a cellular process that engenders fibrosis, by which epithelial cells are transformed into myofibroblasts by losing cell–cell adhesion and by gaining migratory and invasive functions48. As trigger words for “Molecular_function” event, the UMLS ID for mutation was observed most frequently (Table 5e). For “Conversion”, the UMLS ID for ‘Phosphorylation’ was most frequently observed (Table 5f). As trigger words for “Pathway” event, the UMLS IDs for ‘Signal Pathways’ and ‘Metabolic Networks’, which are not specific networks, were observed most frequently (Table 5g). For “Gene_expression”, there are only three UMLS IDs for translation, transcription, and gene expression, among which the ID for translation was by far the most frequently observed (Table 5h).
The event arguments were also analyzed (Tables 6 and 7). Major arguments, ‘Theme’ and ‘Cause’, which are adopted by various event types, tend to take various entities and events (Table 6a,b), whereas ‘atLoc’, which indicates the location at which the corresponding event occurs, takes either “Anatomical_entity” or “Cell” frequently (Table 6c). Regarding the ‘Theme’ argument, the molecular entity “GGPs” is observed most frequently in “Localization”, “Negative_regulation”, “Correlation”, “Molecular_function”, “Conversion”, “Gene_expression”, and “Binding” (Table 6a). Molecular events such as “Molecular_function” and “Gene_expression” were also observed frequently as ‘Theme’ in various events (Table 6a). Regarding ‘Cause’, “Pharmacological_substance”, and “Organic_compound_other”, as well as “GGPs” are also observed frequently in “Positive_regulation”, and “Negative_regulation”.
The frequently observed UMLS IDs were also analyzed for the arguments (Table 7). The UMLS ID for ‘IPF’ was observed most frequently as ‘Theme’ in two events: “Biological_process” and “Correlation” (Table 7a). In comparison with ‘Theme’, the UMLS IDs for various molecules are observed frequently as ‘Cause’ in various events, “Biological_process”, “Regulation”, “Positive_regulation”, “Negative_regulation”, and “Conversion” (Table 7b). It is natural that the UMLS ID for ‘IPF’ was the most frequently observed as ‘disorder’ in various events (Table 7c). It is also natural that the UMLS ID for ‘Lung’ is observed frequently as ‘atLoc’ in various events (Table 7d).
Evaluation results by ten-fold cross-validation
Using ten-fold cross-validation, named entity recognition (NER), entity-linking, event extraction, and relation extraction were conducted to evaluate this corpus. Cross-validation is aimed at evaluating the corpus consistency, and also at examining how much state-of-the-art text-mining systems can address these tasks in the corpus.
Overall F1 scores for entities and event triggers by NER were, respectively, 87.43 and 84.40 (Table 8), which indicates that this corpus can contribute to text-mining for IPF research in terms of NER. However, F1 scores for “Genetic_info”, “Inorganic_compound”, “Cell_component”, and “Binding”, for which the occurrences were very few, are lower than 50.0 (Table 8a). Particularly, the F1 score for “Binding” was zero because the number of occurrences is only eight (Tables 1 and 8a). The F1 scores of NER are correlated with the number of occurrences (Tables 1, 2, and 8) (correlation coefficients were 0.62 for entities and 0.53 for event triggers). Moreover, because a small number of entities and event triggers cannot be distributed equally in all folds in ten-fold cross-validation, some folds contain no such entities and event triggers, which engender zero precision, recall, and F1. Such deviations of the distribution are apparently negatively correlated with the F1 scores. From more specific viewpoints of event triggers, the F1 scores for event triggers of “Regulation” and “Correlation”, 61.96 and 75.26, respectively, are much lower than those of “Positive_regulation” and “Negative_regulation”, 91.61 and 92.35, respectively (Table 8b). Because it is difficult to distinguish event triggers for “Regulation” and “Correlation” from those for “Positive_regulation”, the performance of “Regulation” and “Correlation” might be lower. Regarding IAA measurement, the IAA score for NER of entities and cues shows 79.42, whereas that of event triggers shows 71.31. These IAA scores are lower than the F1 scores for NER by ten-fold cross-validation (87.43 for entities and cues; 84.40 for event triggers) (Table 8).
Results of entity-linking for ten-fold cross-validation are presented in Table 9. As a whole, the performance of entity-linking for entities is good: the F1 score of entity-linking for entities is 68.21 (Table 9a). Because the UMLS IDs for “Genetic_info”, “Negation_cue”, and “Speculation_cue” are not annotated, these data are not included in Table 9a. The F1 scores for “Cell_component” and “Inorganic_compound”, for which the numbers of occurrences were fewer than 30, were lower than 30. The F1 scores of entity-linking for entities correlate with the numbers of occurrences for entities (Tables 1 and 9a) (correlation coefficient, 0.52). However, the F1 score of entity-linking for event triggers is 58.21 (Table 9b), which is lower than that of the entities. The F1 scores for “Regulation”, “Conversion”, “Pathway”, and “Binding” were lower than 30. Particularly, the F1 score for “Binding” was 0.00. Regarding “Conversion”, “Pathway”, and “Binding”, it seems natural that the F1 scores are very low because their occurrences were fewer than 150 (Table 2). The F1 scores of entity-linking for event triggers correlate with the numbers of occurrences for event triggers (Tables 2 and 9b) (correlation coefficient, 0.81), and also with the F1 scores for event triggers in NER (Tables 8b and 9b) (correlation coefficient, 0.73). Regarding the IAA measurement, the IAA score for entity-linking for entities is 72.27, which is lower than that of NER for entities and cues (79.42). However, the IAA score for entity-linking for event triggers is 84.08, which is much higher than that of NER for event triggers (71.31). In contrast to the IAA scores for NER, these IAA scores are higher than the F1 scores for entity-linking by ten-fold cross-validation (68.21 for entities and cues; 58.21 for event triggers) (Table 9).
Results of event extraction and relation extraction, which usually exhibits worse performance than NER in any corpus, are presented in Table 10. The F1 score of event extraction is 45.08: markedly lower than 50 (Table 10a). As a whole, F1 scores of events tend to be lower than 50.0, although those for “Biological_process”, “Cellular_process”, and “Gene_expression” are approximately 60.0, which is higher than the other events (Table 10a). In the MLEE corpus43, the F score for event extraction of anatomical events, which correspond to “Biological_process” and “Cellular_process” in our corpus, is the highest among all the events, suggesting that these events are readily extracted. The F1 scores of the event extraction are not so correlated with the number of occurrences (Tables 2 and 10a) (correlation coefficient, 0.33), but correlated with the F1 scores of event triggers in NER (Tables 8b and 10a) (correlation coefficient, 0.75). However, although the F1 scores of NER event triggers for “Positive_regulation” and “Negative_regulation” are very high (91.61 and 92.35, respectively) (Table 8b), those F1 scores of event extraction are rather low (35.97 and 41.11, respectively) (Table 10a). Generally, the performance of event extraction for such regulation events is lower than those for other events, considering other corpora such as the Cancer Genetics (CG) corpus and the Pathway corpus17,18,19, and the GENIA corpus49. In comparison with the F1 scores of event extraction for the MLEE corpus and the CG corpus using DeepEventMine13,50, the F1 scores of this corpus tend to be lower than these previous corpora, probably due to the larger number of arguments and increased degree of expressions for trigger words. For instance, in the case of “Gene_expression”, F1 score of this corpus showed 59.34, whereas those scores of the MLEE and the CG corpora were 80.80 and 82.64, respectively50. In the case of “Pathway”, F1 score of this corpus showed 54.01, whereas those of the MLEE and the CG corpora were 69.33 and 73.54, respectively50. By introducing a new argument, ‘disorder’, the event structures for this corpus became even more complicated. Moreover, the regulation events often include other events as arguments (‘Theme’ and ‘Cause’) recursively, which might make their extraction challenging19. Thus, it will be necessary to develop a new event extraction system that can extract such complicated events more efficiently and correctly in the future. The IAA score for event extraction is 53.42, which is higher than that for event extraction by ten-fold cross-validation (45.08) (Table 10a). Moreover, the IAA score for event extraction is much lower than any other IAA score. This lower score suggests that event annotation is most difficult to carry out consistently. It also requires more trained annotation skills than any other annotation, such as entities, normalization (ID assignment) and relations, because event structures are the most complicated with event triggers and their relations with several arguments. Because this corpus dataset was annotated by only one annotator (annotator 1), it is largely free of inconsistencies that are unavoidable in a dataset constructed by multiple annotators, especially in terms of event annotation.
The F1 score of event extraction for event modification is 34.24, which is even lower than that of the event extraction above (Table 10b). The F1 score of ‘Negated’ events is 25.64, whereas that of ‘Speculated’ events is 34.92. Regarding ‘Negated’ events, the instances of ‘Negated’ in the gold data are only 93, which can be a reason why its performance is very low. Furthermore, in the other corpora, such as those for Cancer Genetics and Pathway Curation, the event extraction for event modification was apparently challenging, with F1 scores of approximately 3019.
The F1 score of relation extraction is 49.64, also lower than 50, but slightly better than that of event extraction, probably because the relation models are much simpler than the event models. The F1 scores for “Subject_Disorder” and “Disorder_association” are higher than 40, whereas those for “part_of” and “member_of” are lower than 40. The F1 scores of the relation extraction are not so correlated with the number of occurrences (Tables 3 and 10c) (correlation coefficient, 0.23). The related entities for “Subject_Disorder” and “Disorder_association” are very specific, which might make their extraction easier. In contrast, the relations represented by “part_of” and “member_of” are rather complicated, involving various entity types, which might make the extraction more difficult. The IAA score for relation extraction is 76.35, which is much higher than that by ten-fold cross-validation (49.64) (Table 10c).
Novelty and significance of the corpus
To extract and construct a network that is related to the disorder, IPF, entity-linking and annotation data of IPF-related events are necessary. Because many entities are expressed differently, extracted entities should be assigned with IDs so that the same entities can be matched in the networks. Entity-linking in this corpus enables this ID assignment for entities.
Regarding the IPF-related events, those existing corpora cannot provide ‘disorder’-related event data. In this corpus, ‘disorder’-related events have been annotated as indicated in Fig. 1 (Fig. 1e–j). Combined with this corpus, state-of-the-art text-mining system might be able to extract ‘disorder’-related events that are distinguishable from the other ordinary events (Fig. 1a–d) in the near future.
Moreover, this corpus encompasses multiple levels of organisms from molecular level to the whole body level. As an existing corpus for multiple levels of organisms, the MLEE corpus, which has emphasized angiogenesis, the development of new blood vessels, has been reported43. The types of entities and events in our corpus were compared with those of the MLEE corpus (Table 11). Most of the MLEE entities correspond to the entities in our corpus, except for “PROTEIN DOMAIN OR REGION” and “DNA DOMAIN OR REGION”, which are not defined in our corpus (Table 11a). In our corpus, a molecular entity, “DRUG OR COMPOUND”, of the MLEE corpus was subdivided into the three entities, “Inorganic_compound”, “Organic_compound_other”, and “Pharmacological_substance”. In contrast, various anatomical entities of the MLEE corpus, such as “ANATOMICAL SYSTEM”, “ORGAN”, “MULTI-TISSUE STRUCTURE”, and “TISSUE” are integrated into one entity, “Anatomical_entity”, in our corpus. Although preclinical text data were targeted in our corpus, clinical terms, especially for pulmonary disorders, are annotated in “Measurement”, “Entity_property”, and “Method_cue”, which have not been annotated in the MLEE corpus. With these clinical entities, NER and entity-linking can be performed for the clinical literature on lung diseases.
All MLEE events correspond to events in our corpus (Table 11b). At the cellular level, the MLEE corpus has emphasized “CELL PROLIFERATION” and “CELL DIVISION.” In contrast, the wider scope of the cellular events, including EMT, autophagy and cell communication, has been covered in our corpus (Table 5d). At the anatomical level, the MLEE corpus has mainly emphasized “BLOOD VESSEL DEVELOPMENT”, “DEVELOPMENT”; and angiogenesis-related events, such as “GROWTH” “DEATH”, “BREAKDOWN”, and “REMODELING”, whereas fibrosis-related events such as fibrogenesis, fibrosis, and inflammation, have been annotated more intensively in our corpus (Table 5b). In this corpus, molecules involved in inflammation and fibrosis are listed. For example, molecules involved in NFκB signaling and integrin signaling which are related to inflammatory cytokines51. As related to fibrosis, molecules such as TGFβ, surfactant proteins and molecules involved in the Wnt-β catenin signaling are also included51,52. The background of pathological process from inflammation to fibrosis can be understood by discovering the relationships and regulatory relations among these molecules. With these differences from the MLEE corpus, our corpus can emphasize the pulmonary disorder-related events and can facilitate extraction of these events.
Although the reuse of the existing corpora in the annotation and evaluation were not addressed in this study because reuse is beyond the scope of our study, the existing corpora can facilitate improvement of the performance of the disorder-related event extraction by combining our corpus with the existing corpora. We leave this as a subject for future work.
Conclusion
We have presented a new corpus for molecular and cellular mechanisms for a chronic fibrosing interstitial lung disease, idiopathic pulmonary fibrosis (IPF)53. The corpus is expected to be useful to extract IPF pathogenesis mechanisms automatically from huge amounts of scientific texts. We defined entities, events, and relations, annotated a corpus of 150 abstracts, and applied existing state-of-the-art NER and event extraction systems to the corpus. By obtaining timely molecular information from previous reports, we can find the missing links in the previous findings using this corpus combined with the recent text-mining systems. Thus, we will extract molecules related to the acute exacerbation and progressive respiratory failure, or molecules related to inflammation and fibrosis, and furthermore, we will draw their relationship. Moreover, we can find the upstream regulatory molecules of the extracted molecules. We believe that these analyses will help in the search for therapeutic methods. Although this corpus has emphasized IPF, it is applicable to the extraction of information related to other lung diseases, including lung cancer and interstitial pneumonia caused by COVID-19 because some entities and events of this corpus are related also to such diseases.
Data availability
The following datasets are freely available at their respective websites. The corpus for IPF pathogenetic mechanisms: https://ezcatdb.github.io/prism_IPFdata/IPF_corpus/. IAA dataset. Data by annotator 1: https://ezcatdb.github.io/prism_IPFdata/iaa/iaa_1/. Data by annotator 2: https://ezcatdb.github.io/prism_IPFdata/iaa/iaa_2/. Annotation guideline for this work: https://ezcatdb.github.io/prism_IPFdata/AnnotationGuideline_IPFmechanism.pdf.
Abbreviations
- BENNERD:
-
BERT-based exhaustive neural named entity recognition and disambiguation
- BERT:
-
Bidirectional encoder representations from transformers
- BioNLP:
-
Biomedical natural language processing workshop
- COVID-19:
-
Coronavirus disease 2019
- CT:
-
Computed tomography
- CTGF:
-
Connective tissue growth factor
- CUI:
-
Concept unique identifier
- EMT:
-
Epithelial to mesenchymal transition
- EPI:
-
Epigenetic and post-translational modification
- FGF:
-
Fibroblast growth factor
- FVC:
-
Forced vital capacity
- GGPs:
-
Gene and gene products
- GREC:
-
Gene regulation event corpus
- IAA:
-
Inter-annotator agreement
- IPF:
-
Idiopathic pulmonary fibrosis
- MLEE:
-
Multi-level event extraction
- MMLite:
-
MetaMap Lite
- MMP:
-
Matrix metalloproteinase
- mTOR:
-
Mammalian target of rapamycin
- NCI:
-
US National Cancer Institute
- NER:
-
Named entity recognition
- PDGF:
-
Platelet-derived growth factor
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- TGF:
-
Transforming growth factor
- UMLS:
-
Unified medical language system
- VEGF:
-
Vascular endothelial growth factor
References
Raghu, G. et al. An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary fibrosis: Evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 183, 788–824 (2011).
Oda, K. et al. Efficacy of concurrent treatments in idiopathic pulmonary fibrosis patients with a rapid progression of respiratory failure: An analysis of a national administrative database in Japan. BMC Pulm. Med. 16, 91. https://doi.org/10.1186/s12890-016-0253-x (2016).
Nakashima, K. et al. Three cases of sequential treatment with nintedanib following pulsed-dose corticosteroids for acute exacerbation of interstitial lung diseases. Respir. Med. Case Rep. 33, 101385. https://doi.org/10.1016/j.rmcr.2021.101385 (2021).
Noble, P. W. et al. Pirfenidone in patients with idiopathic pulmonary fibrosis (capacity): Two randomised trials. Lancet 377, 1760–1769 (2011).
Richeldi, L. et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N. Engl. J. Med. 370, 2071–2082 (2014).
Schaefer, C. J., Ruhrmund, D. W., Pan, L., Seiwert, S. D. & Kossen, K. Antifibrotic activities of pirfenidone in animal models. Eur. Respir. Rev. 20, 85–97 (2011).
PubMed database. https://pubmed.ncbi.nlm.nih.gov/. (accessed 3 Mar 2022).
McEntyre, J. & Lipman, D. Pubmed: Bridging the information gap. CMAJ 164, 1317–1319 (2001).
Ananiadou, S., Pyysalo, S., Tsujii, J. & Kell, D. B. Event extraction for systems biology by text mining the literature. Trends Biotechnol. 28, 381–390 (2010).
NERsuite. http://nersuite.nlplab.org/index.html. (accessed 15 Mar 2022).
Miwa, M., Saetre, R., Kim, J. D. & Tsujii, J. Event extraction with complex event classification using rich features. J. Bioinform. Comput. Biol. 8, 131–146 (2010).
Miwa, M. & Ananiadou, S. Adaptable, high recall, event extraction system with minimal configuration. BMC Bioinform. 16, S7. https://doi.org/10.1186/1471-2105-16-S10-S7 (2015).
Trieu, H. L. et al. DeepEventMine: end-to-end neural nested event extraction from biomedical texts. Bioinformatics 36, 4910–4917 (2020).
Kim, J. D., Ohta, T., Tateisi, Y. & Tsujii, J. GENIA corpus–semantically annotated corpus for bio-textmining. Bioinformatics 19, i180–i182 (2003).
Kim, J. D., Ohta, T. & Tsujii, J. Corpus annotation for mining biomedical events from literature. BMC Bioinform. 9, 10. https://doi.org/10.1186/1471-2105-9-10 (2008).
Thompson, P., Iqbal, S. A., McNaught, J. & Ananiadou, S. Construction of an annotated corpus to support biomedical information extraction. BMC Bioinform. 10, 349. https://doi.org/10.1186/1471-2105-10-349 (2009).
Cancer Genetics corpus. https://sites.google.com/site/bionlpst2013/tasks/cancer-genetics-cg-task. (accessed 15 Mar 2022).
Pyysalo, S., Ohta, T. & Ananiadou, S. Overview of the cancer genetics (CG) task of bionlp shared task 2013. In Proceedings of the CG, BioNLP Shared Task 2013 Workshop, Association for Computational Linguistics, Sofia, Bulgaria, 58–66. https://www.aclweb.org/anthology/W13-2008.pdf (2013).
Pyysalo, S. et al. Overview of the cancer genetics and pathway curation tasks of BioNLP Shared Task 2013. BMC Bioinform. 16, S2. https://doi.org/10.1186/1471-2105-16-S10-S2 (2015).
Brat rapid annotation tool. https://github.com/nlplab/brat. (accessed 14 Apr 2022).
Stenetorp, P., Pyysalo, S., Topić, G., Ananiadou, S. & Tsujii, J. brat: A web-based tool for NLP-assisted text annotation. In Proceedings of the Demonstrations Session at EACL 2012, Association for Computational Linguistics, Avignon, France, 102–107. https://www.aclweb.org/anthology/E12-2021.pdf (2012).
GENIA meta-knowledge corpus. http://www.nactem.ac.uk/meta-knowledge/. (accessed 15 Mar 2022).
Thompson, P., Nawaz, R., McNaught, J. & Ananiadou, S. Enriching a biomedical event corpus with meta-knowledge annotation. BMC Bioinform. 12, 393. https://doi.org/10.1186/1471-2105-12-393 (2011).
PHAEDRA corpus. http://www.nactem.ac.uk/PHAEDRA/. (accessed 15 Mar 2022).
Thompson, P. et al. Annotation and detection of drug effects in text for pharmacovigilance. J. Cheminform. 10, 37. https://doi.org/10.1186/s13321-018-0290-y (2018).
UMLS database. https://www.nlm.nih.gov/research/umls/licensedcontent/umlsarchives04.html. (accessed 3 Mar 2022).
Bodenreider, O. The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 32, D267–D270 (2004).
Lindberg, D. A., Humphreys, B. L. & McCray, A. T. The unified medical language system. Methods Inf. Med. 32, 281–291 (1993).
Ohta, T., Kim, J. D., Pyysalo, S., Wang, Y. & Tsujii, J. Incorporating genetag-style annotation to GENIA corpus. In Proceedings of BioNLP’09, Association for Computing Machinery. 106–107 https://aclanthology.org/W09-1313.pdf (2009).
Ohta, T., Pyysalo, S., Kim, J. D. & Tsujii, J. A re-evaluation of biomedical named entity-term relations. J. Bioinform. Comput. Biol. 8, 917–928 (2010).
Nawaz, R., Thompson, P. & Ananiadou, S. Negated bio-events: Analysis and identification. BMC Bioinform. 14, 14. https://doi.org/10.1186/1471-2105-14-14 (2013).
MetaMap Lite. https://lhncbc.nlm.nih.gov/ii/tools/MetaMap/run-locally/MetaMapLite.html. (accessed 3 Mar 2022).
Demner-Fushman, D., Rogers, W. J. & Aronson, A. R. MetaMap Lite: An evaluation of a new Java implementation of MetaMap. J. Am. Med. Inform. Assoc. 24, 841–844 (2017).
NCI Metathesaurus. https://ncimeta.nci.nih.gov/ncimbrowser/. (accessed 15 Mar 2022).
Aronson, A. R., Rindflesch, T. C. & Browne, A. C. Exploiting a large thesaurus for information retrieval. Proc. RIAO. 94, 197–216 (1994).
Aronson, A. R. & Lang, F. M. An overview of MetaMap: Historical perspective and recent advances. J. Am. Med. Inform. Assoc. 17, 229–236 (2010).
Annodoc documentation support system. https://spyysalo.github.io/annodoc/. (accessed 15 Mar 2022).
Pyysalo, S. & Ginter, F. Collaborative development of annotation guidelines with application to universal dependencies. SLTC 2014. Swedish Language Technology Conferences, Uppsala, Finland. https://www2.lingfil.uu.se/SLTC2014/abstracts/sltc2014_submission_32.pdf (2014).
IAA dataset. Data by annotator 1: https://ezcatdb.github.io/prism_IPFdata/iaa/iaa_1/ and annotator 2: https://ezcatdb.github.io/prism_IPFdata/iaa/iaa_2/ (2023).
Annotation guideline for this work. https://ezcatdb.github.io/prism_IPFdata/AnnotationGuideline_IPFmechanism.pdf. (2023).
Sohrab, M. G. et al. BENNERD: A neural named entity linking system for COVID-19. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations. Association for Computational Linguistics. 182–188 https://aclanthology.org/2020.emnlp-demos.24.pdf (2020).
Kim, J. D., Ohta, T., Pyysalo, S., Kano, Y. & Tsujii, J. Extracting bio-molecular events from literature–the BioNLP'09 Shared Task. Comput. Intell. 27, 513–540. https://doi.org/10.1111/j.1467-8640.2011.00398.x (2011).
Pyysalo, S. et al. Event extraction across multiple levels of biological organization. Bioinformatics 28, i575–i581 (2012).
Valeyre, D. et al. Sarcoidosis. Lancet 383, 1155–1167 (2014).
Hena, K. M. Sarcoidosis epidemiology: Race matters. Front. Immunol. 11, 537382. https://doi.org/10.3389/fimmu.2020.537382 (2020).
Morimoto, T. et al. Epidemiology of sarcoidosis in Japan. Eur. Respir. J. 31, 372–379 (2008).
Kim, J. D. et al. Overview of BioNLP Shared Task 2011. In Proceedings of the BioNLP 2011 Shared Task, Association for Computational Linguistics, Portland, Oregon, USA, 1–6 https://www.aclweb.org/anthology/W11-1801.pdf (2011).
Rubio, K., Castillo-Negrete, R. & Barreto, G. Non-coding RNAs and nuclear architecture during epithelial–mesenchymal transition in lung cancer and idiopathic pulmonary fibrosis. Cell. Signal. 70, 109593. https://doi.org/10.1016/j.cellsig.2020.109593 (2020).
Kim, J. D. et al. The GENIA event and protein coreference tasks of the BioNLP Shared Task 2011. BMC Bioinform. 13, S1. https://doi.org/10.1186/1471-2105-13-S11-S1 (2012).
Scores by DeepEventMine. https://github.com/aistairc/DeepEventMine (accessed 16 Mar 2023).
Sieber, P. et al. Nf-κB drives epithelial–mesenchymal mechanisms of lung fibrosis in a translational lung cell model. JCI Insight. 8, e154719. https://doi.org/10.1172/jci.insight.154719 (2023).
Wynn, T. A. & Ramalingam, T. R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–1040 (2012).
Corpus for IPF pathogenetic mechanisms. https://ezcatdb.github.io/prism_IPFdata/IPF_corpus/ (2023).
Acknowledgements
This work is based on results obtained from a project commissioned by the Public/Private R&D Investment Strategic Expansion PrograM (PRISM). For experiments, computational resource of AI Bridging Cloud Infrastructure (ABCI) provided by National Institute of Advanced Industrial Science and Technology (AIST) was used.
Funding
This work was supported by the Public/Private R&D Investment Strategic Expansion PrograM (PRISM) in Japan.
Author information
Authors and Affiliations
Contributions
N.N. designed annotation schemes, prepared annotation guideline and datasets, analyzed tendencies of the corpus, and was a major contributor to writing of the manuscript. N.T. performed text-annotation of 120 abstracts and 30 IAA abstracts. M.I. performed text-annotation of 30 IAA abstracts. H.I. contributed to the improvement of guideline by preliminary text-annotation of the IAA abstracts. D.A.K. performed evaluation of the corpus, instructed by M.M. D.A.K. and M.M. were contributors to writing of the manuscript for evaluation methods. M.G.S. prepared the entity-linking system. G.T. performed MetaMap Lite and implemented the brat system. M.N.I. selected abstracts for annotation as the IPF specialist. H.T. designed and supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nagano, N., Tokunaga, N., Ikeda, M. et al. A novel corpus of molecular to higher-order events that facilitates the understanding of the pathogenic mechanisms of idiopathic pulmonary fibrosis. Sci Rep 13, 5986 (2023). https://doi.org/10.1038/s41598-023-32915-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32915-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.