Abstract
Global Developmental Delay/Intellectual disability (ID) is the term used to describe various disorders caused by abnormal brain development and characterized by impairments in cognition, communication, behavior, or motor skills. In the past few years, whole-exome sequencing (WES) has been proven to be a powerful, robust, and scalable approach for candidate gene discoveries in consanguineous populations. In this study, we recruited 215 patients affected with ID from 118 Middle Eastern families. Whole-exome sequencing was completed for 188 individuals. The average age at which WES was completed was 8.5 years. Pathogenic or likely pathogenic variants were detected in 32/118 families (27%). Variants of uncertain significance were seen in 33/118 families (28%). The candidate genes with a possible association with ID were detected in 32/118 (27%) with a total number of 64 affected individuals. These genes are novel, were previously reported in a single family, or cause strikingly different phenotypes with a different mode of inheritance. These genes included: AATK, AP1G2, CAMSAP1, CCDC9B, CNTROB, DNAH14, DNAJB4, DRG1, DTNBP1, EDRF1, EEF1D, EXOC8, EXOSC4, FARSB, FBXO22, FILIP1, INPP4A, P2RX7, PRDM13, PTRHD1, SCN10A, SCYL2, SMG8, SUPV3L1, TACC2, THUMPD1, XPR1, ZFYVE28. During the 5 years of the study and through gene matching databases, several of these genes have now been confirmed as causative of ID. In conclusion, understanding the causes of ID will help understand biological mechanisms, provide precise counseling for affected families, and aid in primary prevention.
Similar content being viewed by others
Introduction
Global Developmental Delay (GDD)/Intellectual disability (ID) represents a group of genetic, phenotypic, and clinically heterogenic disorders that affect approximately 1% of children worldwide1. Significant limitations define GDD or ID in both intellectual functioning and adaptive behavior that originates during brain development. Non-genetic causes such as infections, autoimmunity, and environmental factors are described, but the majority of such disorders have a genetic basis2.
Hundreds of genes are thought to be involved in the etiology of ID3. The list of ID genes has expanded and according to the SysNDD database, there are now 2841 primary and candidate human ID genes4. In the last decade, advances in genetic technologies such as next-generation sequencing (NGS) have revolutionized clinical practice in medical genetics, aided clinical diagnosis, and proved to be very effective in discovering an ever-increasing number of ID-related genes. They have also enabled deciphering the ID’s heterogeneous genetic mechanisms5.
Whole-Exome Sequencing (WES), as a clinical diagnostic test, has a success rate of about 30–40%6. The diagnostic yield of chromosomal microarray in children with no underlying cause of their ID is around 15 to 20%7. In trio-based WES done in groups of children with severe ID, the yield ranged from 13 to 35%8. In contrast, exome sequencing in samples from consanguineous families with various ID-associated phenotypes has produced a high yield. For example, in several studies of ID from the Middle East, NGS yield ranged from 37 to 90%, depending on the patients’ cohorts, the study design, and the classification of the variants9,10,11,12,13,14,15.
This study reports on the diagnostic yield and candidate genes of a genetic study of intellectual disability in 118 Omani families. The study period extended over five years and included 215 affected individuals. The diagnostic yield when considering both pathogenic and uncertain variants was 55%. Candidate genes with a possible association with ID phenotype seen in this cohort were detected in 32/118 (27%) with a total number of 64 affected individuals. Understanding ID causes will provide precise counseling for affected families and aid in primary prevention. Also, the costs of unnecessary investigations will be spared, with fewer diagnostic odysseys. The paper also discusses the pitfalls and challenges of candidate gene discovery in a consanguineous population.
Materials and methods
Human subjects
The Medical Research Ethics Committee approved the study of Sultan Qaboos University (SQU MREC#1362). Informed written consent was obtained from all participants or their guardians. All methods were performed in accordance with the relevant guidelines and regulations and in accordance with the declaration of Helsinki. The target patients included in this study presented with global developmental delay or intellectual disability, all assessed clinically by medical geneticists (detailed methods in SUPP_S1). Severe phenotypes causing death within the neonatal or early infantile period were also included as a neurological phenotype was evident, such as seizures, hypotonia, or brain malformations. Families with a likely autosomal recessive pattern of inheritance were selected. Patients with known molecular diagnoses at the time of recruitment were excluded. The study and exome data analysis were carried out over 5 years, between 2016 and 2021.
Whole exome sequencing and variant interpretation
Whole-exome sequencing analysis was performed for all affected individuals where samples were available. A detailed methodology is presented in the supplemental data (SUPP_S1). In brief, the method used hybrid capture technology (Agilent SureSelect Human All-exons-V6 or V7) for exome enrichment and capture. Illumina technology (Hiseq2500, Hiseq4000, or NovaSeq6000) of 150 bp paired-end, at 150-200X coverage, was used for sequencing. The reads were mapped against UCSC GRCh37/hg19 or GRCh38/hg38. Filtering and variant prioritization were analyzed using an in-house pipeline (SUPP_S1). Variant filtration was performed to keep only novel or rare variants (≤ 1%). Public databases such as 1000 Genomes, Exome Variant Server, and GnomAD were used for alleles frequencies. For filtration of common variants against the Middle Eastern population, the Greater Middle East (GME) and variome database “al mena” that comprises data of 2497 samples was used16,17. Our in-house population-specific exomes database, which contains data of 1564 WES, was also used. During the filtration process, the phenotype and mode of inheritance were both considered. Any potential variants identified after prioritization were further confirmed by Sanger sequencing. Members included in the segregation analysis ranged from 3 to 12 members of each family, depending on the DNA availability. Most of the segregation was performed for the parents and siblings alongside the index patient. When a definitive cause was not possible or when a candidate gene was considered, further analysis of copy number variants was performed on exome data using ExomeDepth18 (SUPP_S1).
Classification of variants was based on the published ACMG guideline19,20. Pathogenic or likely pathogenic variants in known disease-causing genes which could be linked to the reported phenotypes of the affected patients were categorized as disease-causing variants. The second category was for the variants in known disease-causing genes that overlapped with the patient’s phenotype, and these were considered possible disease-causing variants. These were rare and damaging variants of uncertain significance (VUS). Variants in candidate genes, which were predicted to be deleterious and found in genes not previously confirmed to be implicated in human disease, formed the third category. These genes are novel; they were previously reported in a single family or cause strikingly different phenotypes with different modes of inheritance. Supporting data for candidate genes included variants within a shared autozygosity area. The variant is of high or moderate impact; the population frequency supports genic intolerance to such variants, and in-silico prediction tools indicate a damaging effect. Data for gene function and network, gene expression, and animal models were also considered.
Results
Whole-exome sequence analysis was performed for 188 individuals representing 118 characterized families with a total number of 215 affected individuals. Of the 118 families included, 93 (78.8%) had a family history of one or more affected individuals in addition to the index patient, all with a similar phenotype. The age range of the affected individuals, at first clinical assessment, was from birth to 34 years old. The average age at which WES was completed was 8.5 years, and children below five years of age, represented 30% of the affected individuals when WES was completed. Males represented 57.2% of the studied group (123 Males:92 Females). The rate of consanguineous marriages within the included families was 91%. The affected patients exhibited diverse phenotypes, including global developmental delay, seizures, brain malformations, microcephaly, facial dysmorphism, and other systemic manifestations (Table 1 and Supplementary SUPP_Table1).
A total of 420 members’ DNA samples were available for WES or Sanger sequencing for segregation analysis. These included healthy or affected members. However, DNA samples were not available for analysis in 22 out of the 215 affected individuals. Sanger sequencing was used to confirm the variant and phenotype-genotype segregation in all candidate variants. Only variants that were confirmed and segregated with the phenotype are reported.
Variants in previously known and described ID genes were seen in 65/118 families (55%). Following the ACMG guidelines of variant classification, pathogenic (P) or likely pathogenic (LP) variants were detected in 32/118 families (27%). Variants of uncertain significance were seen in 33/118 families (28%). The majority of these two groups showed homozygous variants (51/65; 78.5%). These variants are rare; they explain the disease manifestations, are predicted to be damaging, are confirmed by Sanger sequencing, and are segregated with the phenotype (All listed in SUPP_Table1).
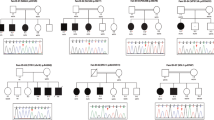
Candidate genes with a possible association with ID phenotype seen in this cohort were detected in 32/118 (27%) with a total number of affected individuals of 64 (Table 1). These candidate genes were selected according to rarity and absence in homozygosity status in local control exomes or public databases. The impact of the variants is predicated damaging. The expression patterns or mouse models supported an association with neurological dysfunction. Importantly, Sanger sequencing confirmed segregation for all variants in candidate genes in up to 3 generations in the family pedigrees (Fig. 1). The total number of candidate genes identified was 28. Table 1 shows detailed findings for the candidate genes.
Twenty-one families (18%) with 40 affected individuals remain unsolved despite multiple molecular tests, including WES, chromosomal microarray, and Fragile X where applicable. There is a trend of milder phenotypes and usually non-syndromic intellectual disabilities in unresolved cases. However, we can not draw a firm conclusion because of the small numbers.
Discussion
Global Developmental Delay (GDD)/Intellectual disability (ID) represents a group of genetic, phenotypic, and clinically heterogenic disorders that affect approximately 1% of children worldwide. This study presents the results of 188 exome analyses representing 118 consanguineous Omani families. This cohort included 215 affected individuals with intellectual disabilities, including global developmental delay, seizures, brain malformations, microcephaly, facial dysmorphism, and other systemic manifestations. Overall 82% were found to have a possible explanation. Specifically, 55% had variants in previously described and known genes (P/LP or VUS) and 27% in possible candidate genes.
With the enrichment for consanguineous families (91%), it was not surprising that the majority (85.5%) of the overall variants in the three groups were homozygous. The consanguinity rate is high due to the preference to include families with autosomal recessive phenotypes and multiple affected individuals. This study detected pathogenic (P) or likely pathogenic (LP) variants in 32 families, making the diagnostic rate of the study 27%. The main goal of this study was to recruit unsolved cases. However, the diagnostic rate observed was higher than anticipated. This can be explained by frequent reanalysis of exome data during the last five years of the study, thus enabling newly published genes to be detected. Also, VUSs, including non-coding variants, were selected with further evidence of pathogenicity becoming available. Also, during the study's initial phase, some families did not have access to clinical exome sequencing and thus were channeled to the research exome.
In a large-scale exome sequencing study, Monies and colleagues15 reported the yield of exome sequencing on 2219 families from Saudi Arabia. The overall diagnostic yield of exome sequencing based on cases with confirmed pathogenic or likely pathogenic variants was 43.3%. However, if considering variants of unknown significance (VUS) that are in an established disease-related gene or candidate genes with compelling biological candidacy were considered, the yield rate would be 73%. The high throughput design of this study led to the discovery of 236 genes that have no established OMIM phenotypes and were proposed as candidate genes. The negative results (unsolved cases) accounted for 27% of the total.
The total number of candidate genes for intellectual disability identified in this study was 28. During the course of this study, Gene Matcher21 was used to provide further evidence of association with the phenotype. Through this study and in collaboration with the scientifc community, several of these genes have been successfully confirmed to cause intellectual disability (Table 1). One interesting shared candidate variant within the XPR1 gene was identified in multiple affected individuals from four apparently unrelated families (Table 1). These families come from different geographical areas of Oman. However, haplotype analysis using exome data indicated that they all shared the same haplotype (Data not shown). The XPR1 protein functions to mediate phosphate export from the cell as well as binding inositol hexakisphosphate and related inositol polyphosphates, which are key intracellular signaling molecules22. Mutations in the XPR1 gene are known to be associated with the dominant condition of idiopathic basal ganglia calcification-6; OMIM 61641323. The earliest age of onset for this condition is in the third and fourth decade of life, with symptoms of cerebrovascular insufficiency associated with movement disorders, cognitive decline and psychiatric symptoms. Our patients’ phenotype is strikingly different; we detected biallelic XPR1 variants and apparently healthy parents. The phenotype included variable signs of neonatal pulmonary hypertension, cardiomyopathy, serum hypophosphatemia, chronic lung disease requiring oxygen therapy, severe developmental delay, and brain basal ganglia calcification. Further functional characterization for these variants has commenced.
Whole-genome sequencing (WGS) analysis can cover up to 98% of the human genome, whereas WES only covers about 95% of the coding regions and only 1–2% of the genome. WES has a lower cost per sample than WGS, a greater depth of coverage in target regions, lesser storage requirements, and easier data analysis24. It is, however, worth highlighting the pitfalls and challenges that can occur when WES analysis is performed. WES is a high-throughput, complex technique with potential pitfalls at every step. These pitfalls and the consequential missing of the molecular diagnosis in exome sequencing and analysis is a recognized phenomenon25. This phenomenon could be caused by various factors, including technological limitations in variant detection, a lack of enrolling additional family members, many variants identified for probands, or the causal variant being located outside of the coding regions25. Pitfalls related to WES analysis can be categorized into three main groups (Table 2).
The first group of pitfalls consists of those which are sequence-related. Large rearrangements or complex structural variants are one example. Structural variants are genomic rearrangements larger than 50 bp in size, and they account for about 1% of the variation in human genomes26. Complex structural variants have been shown to contribute to human genomic variation and to cause Mendelian disease27. Unfortunately, these cannot be identified easily by WES. However, multiple pipelines for CNV analysis are available. In this study and using ExomeDepth, it was possible to detect CNVs in a homozygosity state as a cause of the phenotype (Families 11MS8800 and 10DH12500). Mitochondrial mutations are other causative factors that WES cannot detect. Other factors also include mosaicism, abnormal methylation, and uniparental disomy.
Other causes of missing variants in WES include decreased coverage, locus-specific features such as GC-rich regions, and sequencing biases28,29. Difficulty in the alignment of indels (insertions/deletions) larger than 20–50 nucleotides long is one of the limitations, which is likely to be the reason for WES missing such variants30. Incomplete human genome annotation and sequence might also affect the accuracy of variant mapping and annotation. For instance, an intronic variant could be located in an unannotated exon31. Another potential cause is the high sequence similarity between pseudogenes and their corresponding functional genes32.
The second group to consider is pitfalls due to annotation and prioritization errors. Annotation and prioritization steps are used in WES analyses to reduce thousands of variants to a few candidates. During the filtering process, all annotations except for the “canonical” transcripts (i.e., the longest transcript of a gene) are initially ignored. Remarkably, pathogenic variants can be missed if alternative transcripts are not fully considered33. Splicing is thought to be involved in 15–30% of all inherited disease variants34. Despite advances in exome capture methods or machine learning for detecting variants that affect splicing, accurate detections of deep intronic variants remain limited and this could be the reason for missing splicing variants. In our cohort, intronic variants that were likely to affect splicing were detected in 19/118 families (16.1%) of which 8 were in non-canonical splicing sites.
Databases like OMIM and HGMD are used to find gene-disease and variant–disease associations in the literature. Variants or genes listed in these databases would be flagged as potentially disease-causing35. Nevertheless, a reason for an initial false-negative result is that such variants or disease databases have not been kept up to date. A recent study by Bruel and colleagues illustrated this issue in a study cohort of 313 individuals36. Likewise, when considering deleterious variants, those with high population prevalence might be filtered out. Penetrance of the disease might be influenced by numerous factors, including other hidden rare variants, family history, inheritance, additional medical problems, and ethnic background.
Synonymous variants, which are known as ‘silent’ variants, represent almost 50% of the variant list identified by WES. Filtering out synonymous variants reduces the variants’ list because they are assumed to be benign. However, our increased knowledge about the relationship between genetic variants and disease has shown that synonymous variants play a significant role in human disease risk and other complex traits, including variants that affect splicing37. Indeed, a recent study challenged the concept that synonymous variants are neutral. In their yeast study, Shen and colleagues showed a strong non-neutrality of most synonymous mutations38. If this holds true for other genes and organisms, then numerous biological conclusions, including disease causation about synonymous mutation, would require re-examination. An example in our cohort is the family 16SS2600, where GPT2 (p.Gly245Gly) was initially missed and flagged as a silent variant.
All labs encounter pitfalls related to clinical factors and phenotypes. As a result, a negative WES result must be interpreted in the context of the patient’s clinical history to determine whether reevaluation or further testing is necessary. For example, within our cohort (family 10MS6600), two related families with multiple affected individuals were enrolled as having the same phenotype and WES analysis was initially negative. However, after a detailed clinical reevaluation and exome reanalysis, the results showed that two different diseases were possibly running in the family. Some of the affected members were indeed found to harbor a deep intronic variant in the PGAP3, which was recently reported to cause hyperphosphatasia with mental retardation syndrome type 4 (OMIM 615,716). Another example is family 10MS16500, demonstrating that multiple individuals carrying two or more different diseases can complicate the phenotype. Similarly, the family (10DF10800) with multiple affected individuals presented with developmental delay, congenital cataracts, and bilateral sensorineural hearing loss. WES analysis identified two different variants for two different syndromes in two genes, one of which is novel as the underlying cause39. Another aspect is the mode of inheritance in which conditions are known to be autosomal dominant but manifest as autosomal recessive. Monies and colleagues reported many examples of genes or diseases inherited as both AD and AR15.
Conclusion
In conclusion, using WES to identify the novel causes of human disease has changed the research landscape of genetic and neurodevelopmental disorders. Although WES is comprehensive technology, its limitations must be considered when negative results are obtained. The pitfalls of WES can potentially reduce the effectiveness of this technique in biological and medical research as well as in clinical settings. Finally, it is worth emphasizing that identifying a likely candidate gene is often just the start of a long process to confirm the variant’s pathogenicity.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and Supplementary material. Further derived data are available from the corresponding author upon reasonable request. The datasets generated and/or analysed during the current study are available in the [Clinvar] repository, [https://www.ncbi.nlm.nih.gov/clinvar/; submission number SCV002574702 to SCV002574743].
References
Maulik, P. K., Mascarenhas, M. N., Mathers, C. D., Dua, T. & Saxena, S. Prevalence of intellectual disability: A meta-analysis of population-based studies. Res. Dev. Disabil. 32, 419–436 (2011).
Kvarnung, M. & Nordgren, A. Intellectual disability & rare disorders: A diagnostic challenge. Adv. Exp. Med. Biol. 1031, 39–54 (2017).
Van Bokhoven, H. Genetic and epigenetic networks in intellectual disabilities. Annu. Rev. Genet. 45, 81–104 (2011).
Kochinke, K. et al. Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am. J. Hum. Genet. 98, 149–164 (2016).
Heyne, H. O. et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 50, 1048–1053 (2018).
Farwell, K. D. et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: Results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 17, 578–586 (2015).
D’Arrigo, S. et al. The diagnostic yield of array comparative genomic hybridization is high regardless of severity of intellectual disability/developmental delay in children. J. Child Neurol. 31, 691–699 (2016).
Vissers, L. E. L. M., Gilissen, C. & Veltman, J. A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 17, 9–18 (2016).
Yavarna, T. et al. High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Hum. Genet. 134, 967–980 (2015).
Charng, W. L. et al. Exome sequencing in mostly consanguineous Arab families with neurologic disease provides a high potential molecular diagnosis rate. BMC Med. Genomics 9, 1–14 (2016).
Megahed, H. et al. Utility of whole exome sequencing for the early diagnosis of pediatric-onset cerebellar atrophy associated with developmental delay in an inbred population. Orphanet J. Rare Dis. 11, 57 (2016).
Anazi, S. et al. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry 22, 615–624 (2017).
Reuter, M. S. et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiat. 74, 293–299 (2017).
Santos-Cortez, R. L. P. et al. Novel candidate genes and variants underlying autosomal recessive neurodevelopmental disorders with intellectual disability. Hum. Genet. 137, 735–752 (2018).
Monies, D. et al. Lessons learned from large-scale, first-tier clinical exome sequencing in a highly consanguineous population. Am. J. Hum. Genet. 104, 1182–1201 (2019).
Özçelik, T. & Onat, O. E. Genomic landscape of the Greater Middle East. Nat. Genet. 48, 978–979 (2016).
Koshy, R., Ranawat, A. & Scaria, V. Al mena: A comprehensive resource of human genetic variants integrating genomes and exomes from Arab, Middle Eastern and North African populations. J. Hum. Genet. 62, 889–894 (2017).
Plagnol, V. et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 28, 2747–2754 (2012).
Harrison, S. M., Biesecker, L. G. & Rehm, H. L. Overview of specifications to the ACMG/AMP variant interpretation guidelines. Curr. Protoc. Hum. Genet. 103, e93 (2019).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17, 405–424 (2015).
Sobreira, N., Schiettecatte, F., Valle, D. & Hamosh, A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928–930 (2015).
Yao, X. P. et al. Analysis of gene expression and functional characterization of XPR1: A pathogenic gene for primary familial brain calcification. Cell Tissue Res. 370, 267–273 (2017).
Legati, A. et al. Mutations in XPR1 cause primary familial brain calcification associated with altered phosphate export. Nat. Genet. 47, 579–581 (2015).
Alfares, A. et al. Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet. Med. 20, 1328–1333 (2018).
Corominas, J. et al. Clinical exome sequencing—Mistakes and caveats. Hum. Mutat. https://doi.org/10.1002/humu.24360 (2022).
Tattini, L., D’Aurizio, R. & Magi, A. Detection of genomic structural variants from next-generation sequencing data. Front. Bioeng. Biotechnol. 3, 1–8 (2015).
Sanchis-Juan, A. et al. Complex structural variants in Mendelian disorders: identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 10, 1–10 (2018).
Ross, M. G. et al. Characterizing and measuring bias in sequence data. Genome Biol. 14, R51 (2013).
Lelieveld, S. H., Spielmann, M., Mundlos, S., Veltman, J. A. & Gilissen, C. Comparison of exome and genome sequencing technologies for the complete capture of protein-coding regions. Hum. Mutat. 36, 815–822 (2015).
Pena, L. D. M. et al. Looking beyond the exome: A phenotype-first approach to molecular diagnostic resolution in rare and undiagnosed diseases. Genet. Med. 20, 464–469 (2018).
Krebs, C. E. et al. The Sac1 domain of SYNJ1 identified mutated in a family with early-onset progressive Parkinsonism with generalized seizures. Hum. Mutat. 34, 1200–1207 (2013).
Pink, R. C. et al. Pseudogenes: Pseudo-functional or key regulators in health and diseasě. RNA 17, 792–798 (2011).
Bodian, D. L., Kothiyal, P. & Hauser, N. S. Pitfalls of clinical exome and gene panel testing: alternative transcripts. Genet. Med. 21, 1240–1245 (2019).
Mertes, C. et al. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat. Commun. 12, 1–13 (2021).
Kesselheim, A., Ashton, E. & Bockenhauer, D. Potential and pitfalls in the genetic diagnosis of kidney diseases. Clin. Kidney J. 10, 581–585 (2017).
Bruel, A. L. et al. Increased diagnostic and new genes identification outcome using research reanalysis of singleton exome sequencing. Eur. J. Hum. Genet. 27, 1519–1531 (2019).
Sauna, Z. E. & Kimchi-Sarfaty, C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 12, 683–691 (2011).
Shen, X., Song, S., Li, C. & Zhang, J. Synonymous mutations in representative yeast genes are mostly strongly non-neutral. Nature 606, 725–731 (2022).
Alzahrani, F. et al. Recessive, deleterious variants in SMG8 expand the role of nonsense-mediated decay in developmental disorders in humans. Am. J. Hum. Genet. 107, 1178–1185 (2020).
Baker, S. J. et al. Characterization of an alternatively spliced AATYK mRNA: Expression pattern of AATYK in the brain and neuronal cells. Oncogene 20, 1015–1021 (2001).
Barik, S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 36, 5232–5241 (2008).
Lewin, D. A. et al. Cloning, expression, and localization of a novel γ-adaptin-like molecule. FEBS Lett. 435, 263–268 (1998).
Zizioli, D. et al. γ2 and γ1AP-1 complexes: Different essential functions and regulatory mechanisms in clathrin-dependent protein sorting. Eur. J. Cell Biol. 96, 356–368 (2017).
Usmani, M. A. et al. De novo and bi-allelic variants in AP1G1 cause neurodevelopmental disorder with developmental delay, intellectual disability, and epilepsy. Am. J. Hum. Genet. 108, 1330–1341 (2021).
Jiang, K. et al. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell 28, 295–309 (2014).
King, M. D. A. et al. A conserved sequence in calmodulin regulated spectrin-associated protein 1 links its interaction with spectrin and calmodulin to neurite outgrowth. J. Neurochem. 128, 391–402 (2014).
Zou, C. et al. Centrobin: A novel daughter centriole-associated protein that is required for centriole duplication. J. Cell Biol. 171, 437–445 (2005).
Sakamoto, K. et al. Ependymal ciliary motion and their role in congenital hydrocephalus. Child’s Nerv. Syst. 37, 3355–3364 (2021).
Shamseldin, H. E. et al. Identification of embryonic lethal genes in humans by autozygosity mapping and exome sequencing in consanguineous families. Genome Biol. 16, 1–7 (2015).
Li, J. et al. DNAH14 variants are associated with neurodevelopmental disorders. Hum. Mutat. 43, 940–949 (2022).
Qiu, X. B., Shao, Y. M., Miao, S. & Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570 (2006).
Ancevska-Taneva, N., Onoprishvili, I., Andria, M. L., Hiller, J. M. & Simon, E. J. A member of the heat shock protein 40 family, hlj1, binds to the carboxyl tail of the human mu opioid receptor. Brain Res. 1081, 28–33 (2006).
Lei, J. X., Cassone, C. G., Luebbert, C. & Liu, Q. Y. A novel neuron-enriched protein SDIM1 is down regulated in Alzheimer’s brains and attenuates cell death induced by DNAJB4 over-expression in neuro-progenitor cells. Mol. Neurodegener. 6, 9 (2011).
Schellhaus, A. K. et al. Developmentally regulated GTP binding protein 1 (DRG1) controls microtubule dynamics. Sci. Rep. 7, 9996 (2017).
Sazuka, T. et al. Expression of DRG during murine embryonic development. Biochem. Biophys. Res. Commun. 189, 371–377 (1992).
Kumar, S., Iwao, M., Yamagishi, T., Noda, M. & Asashima, M. Expression of GTP-binding protein gene drg during Xenopus laevis development. Int. J. Dev. Biol. 37, 539–546 (1993).
Tang, T. T. T. et al. Dysbindin regulates hippocampal LTP by controlling NMDA receptor surface expression. Proc. Natl. Acad. Sci. U. S. A. 106, 21395–21400 (2009).
Cheah, S. Y., Lawford, B. R., Young, R. M., Morris, C. P. & Voisey, J. Dysbindin (DTNBP1) variants are associated with hallucinations in schizophrenia. Eur. Psychiatry 30, 486–491 (2015).
Maes, M. S. et al. Schizophrenia-associated gene dysbindin-1 and tardive dyskinesia. Drug Dev. Res. 82, 678–684 (2021).
Wang, D., Li, Y. & Shen, B. A novel erythroid differentiation related gene EDRF1 upregulating globin gene expression in HEL cells. Chin. Med. J. (Engl) 115, 1701–1705 (2002).
Kaitsuka, T. & Matsushita, M. Regulation of translation factor EEF1D gene function by alternative splicing. Int. J. Mol. Sci. 16, 3970–3979 (2015).
McLachlan, F., Sires, A. M. & Abbott, C. M. The role of translation elongation factor eEF1 subunits in neurodevelopmental disorders. Hum. Mutat. 40, 131–141 (2019).
Coulter, M. E. et al. Regulation of human cerebral cortical development by EXOC7 and EXOC8, components of the exocyst complex, and roles in neural progenitor cell proliferation and survival. Genet. Med. 22, 1040–1050 (2020).
Ullah, A. et al. A novel nonsense variant in EXOC8 underlies a neurodevelopmental disorder. Neurogenetics 23, 203–212 (2022).
Sato, R. et al. Novel biallelic mutations in the PNPT1 gene encoding a mitochondrial-RNA-import protein PNPase cause delayed myelination. Clin. Genet. 93, 242–247 (2018).
Van Dijk, E. L., Schilders, G. & Pruijn, G. J. M. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA 13, 1027–1035 (2007).
dos Santos, R. F. et al. Major 3′–5′ Exoribonucleases in the metabolism of coding and non-coding RNA. Prog. Mol. Biol. Transl. Sci. 159, 101–155 (2018).
Matilainen, S. et al. Defective mitochondrial RNA processing due to PNPT1 variants causes Leigh syndrome. Hum. Mol. Genet. 26, 3352–3361 (2017).
Zadjali, F. et al. Homozygosity for FARSB mutation leads to Phe-tRNA synthetase-related disease of growth restriction, brain calcification, and interstitial lung disease. Hum. Mutat. 39, 1355–1359 (2018).
Xu, Z. et al. Bi-allelic mutations in Phe-tRNA synthetase associated with a multi-system pulmonary disease support non-translational function. Am. J. Hum. Genet. 103, 100–114 (2018).
He, Y. et al. Circular RNA circ_0006282 contributes to the progression of gastric cancer by sponging miR-155 to upregulate the expression of FBXO22. Onco. Targets. Ther. 13, 1001–1010 (2020).
Zheng, X., Yu, S., Xue, Y. & Yan, F. FBXO22, ubiquitination degradation of PHLPP1, ameliorates rotenone induced neurotoxicity by activating AKT pathway. Toxicol. Lett. 350, 1–9 (2021).
Nagano, T. et al. Filamin A-interacting protein (FILIP) regulates cortical cell migration out of the ventricular zone. Nat. Cell Biol. 4, 495–501 (2002).
Kwon, M. et al. Functional characterization of filamin a interacting protein 1–like, a novel candidate for antivascular cancer therapy. Cancer Res. 68, 7332–7341 (2008).
Ivetac, I. et al. The type Iα inositol Polyphosphate 4-Phosphatase generates and terminates phosphoinositide 3-kinase signals on endosomes and the plasma membrane. Mol. Biol. Cell 16, 2218–2233 (2005).
Banihashemi, S., Tahmasebi-Birgani, M., Mohammadiasl, J. & Hajjari, M. Whole exome sequencing identified a novel nonsense INPP4A mutation in a family with intellectual disability. Eur. J. Med. Genet. 63(4), 103846 (2020).
Gu, B. J. et al. A rare functional haplotype of the P2RX4 and P2RX7 genes leads to loss of innate phagocytosis and confers increased risk of age-related macular degeneration. FASEB J. 27, 1479–1487 (2013).
Chen, Y. H., Lin, R. R. & Tao, Q. Q. The role of P2X7R in neuroinflammation and implications in Alzheimer’s disease. Life Sci. 271, 119187 (2021).
Chang, J. C. et al. Prdm13 mediates the balance of inhibitory and excitatory neurons in somatosensory circuits. Dev. Cell 25, 182–195 (2013).
Coolen, M. et al. Recessive PRDM13 mutations cause fatal perinatal brainstem dysfunction with cerebellar hypoplasia and disrupt Purkinje cell differentiation. Am. J. Hum. Genet. 109, 909–927 (2022).
Kuipers, D. J. S. et al. PTRHD1 Loss-of-function mutation in an african family with juvenile-onset Parkinsonism and intellectual disability. Mov. Disord. 33, 1814–1819 (2018).
Khodadadi, H. et al. PTRHD1 (C2orf79) mutations lead to autosomal-recessive intellectual disability and parkinsonism. Mov. Disord. 32, 287–291 (2017).
Al-Kasbi, G. et al. Biallelic PTRHD1 frameshift variants associated with intellectual disability, spasticity, and parkinsonism. Mov. Disord. Clin. Pract. 8, 1253–1257 (2021).
Rabert, D. K. et al. A tetrodotoxin-resistant voltage-gated sodium channel from human dorsal root ganglia, hPN3/SCN10A. Pain 78, 107–114 (1998).
Chilton, I. et al. De novo heterozygous missense and loss-of-function variants in CDC42BPB are associated with a neurodevelopmental phenotype. Am. J. Med. Genet. Part A 182, 962–973 (2020).
Kambouris, M. et al. Biallelic SCN10A mutations in neuromuscular disease and epileptic encephalopathy. Ann. Clin. Transl. Neurol. 4, 26–35 (2016).
Gingras, S. et al. SCYL2 protects CA3 pyramidal neurons from excitotoxicity during functional maturation of the mouse hippocampus. J. Neurosci. 35, 10510–10522 (2015).
Seidahmed, M. Z. et al. Recessive mutations in SCYL2 cause a novel syndromic form of arthrogryposis in humans. Hum. Genet. 139, (2020).
van Esveld, S. L. et al. Mitochondrial RNA processing defect caused by a SUPV3L1 mutation in two siblings with a novel neurodegenerative syndrome. J. Inherit. Metab. Dis. 45, 292–307 (2022).
Rutherford, E. L. et al. Xenopus TACC2 is a microtubule plus end-tracking protein that can promote microtubule polymerization during embryonic development. Mol. Biol. Cell 27, 3013–3020 (2016).
Takayama, K. I. et al. TACC2 is an androgen-responsive cell cycle regulator promoting androgen-mediated and castration-resistant growth of prostate cancer. Mol. Endocrinol. 26, 748–761 (2012).
Sharma, S. et al. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 43, 2242–2258 (2015).
Broly, M. et al. THUMPD1 bi-allelic variants cause loss of tRNA acetylation and a syndromic neurodevelopmental disorder. Am. J. Hum. Genet. 109, 587–600 (2022).
Funding
This work was supported by Sultan Qaboos University; study code: SR/MED/GENT/16/01.
Author information
Authors and Affiliations
Contributions
(1) Research project: A. Conception, B. Organization, C. Execution; (2) Experiments: A. Design, B. Execution, C. Review and Critique; (3) Manuscript: A. Writing of the first draft, B. Review and Critique. (4) A. patient care, B. Clinical Phenotype characterization. G.K.:1A, 1B, 1C, 2A, 2B, 3A F.M., A.K., N.H., K.T., A.S., A.F., W.M., A.A.: 1A, 3A, 4A, 4B ZB, K.K.: 4A, 4B SA: 2B, 2C, 3B F.Z.: 1A, 1B, 3C S.Y.: 1A, 1B, 1C, 2A, 2C, 3A, 3B A.M.: 1A, 1B, 1C, 2A, 2B, 2C, 3A, 3B. 4A, 4B .
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Al-Kasbi, G., Al-Murshedi, F., Al-Kindi, A. et al. The diagnostic yield, candidate genes, and pitfalls for a genetic study of intellectual disability in 118 middle eastern families. Sci Rep 12, 18862 (2022). https://doi.org/10.1038/s41598-022-22036-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22036-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.