Abstract
In a staged anaerobic fluidized-bed ceramic membrane bioreactor, metagenomic and metatranscriptomic analyses were performed to decipher the microbial interactions on the granular activated carbon. Metagenome bins, representing the predominating microbes in the bioreactor: syntrophic propionate-oxidizing bacteria (SPOB), acetoclastic Methanothrix concilii, and exoelectrogenic Geobacter lovleyi, were successfully recovered for the reconstruction and analysis of metabolic pathways involved in the transformation of fatty acids to methane. In particular, SPOB degraded propionate into acetate, which was further converted into methane and CO2 by M. concilii via the acetoclastic methanogenesis. Concurrently, G. lovleyi oxidized acetate into CO2, releasing electrons into the extracellular environment. By accepting these electrons through direct interspecies electron transfer (DIET), M. concilii was capable of performing CO2 reduction for further methane formation. Most notably, an alternative RuBisCO-mediated CO2 reduction (the reductive hexulose-phosphate (RHP) pathway) is transcriptionally-active in M. concilii. This RHP pathway enables M. concilii dominance and energy gain by carbon fixation and methanogenesis, respectively via a methyl-H4MPT intermediate, constituting the third methanogenesis route. The complete acetate reduction (2 mole methane formation/1 mole acetate consumption), coupling of acetoclastic methanogenesis and two CO2 reduction pathways, are thermodynamically favorable even under very low substrate condition (down to to 10−5 M level). Such tight interactions via both mediated and direct interspecies electron transfer (MIET and DIET), induced by the conductive GAC promote the overall efficiency of bioenergy processes.
Similar content being viewed by others
Introduction
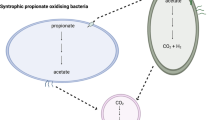
Energy recovery in the form of methane with sewage is of great interest1. Methanogenesis is accomplished by the syntrophic microbial interactions to achieve interspecies electron transfer (IET). Traditionally, IET was thought to be accomplished by syntrophs and methanogens through diffusive carriers (e.g. acetate, hydrogen and formate). This form of mediated IET (MIET) is constrained by the physical distance between syntrophs and methanogens, and the diffusion rate of electron carriers, creating a metabolic bottleneck2. However, there is growing evidence of an alternative direct interspecies electron transfer (DIET), which could overcome this bottleneck and enhance methane production rate3,4. DIET occurs in the presence of exoelectrogens, which are capable of shuttling electrons exogenously to methanogens through conductive pili or surfaces3,5. It is the predominant mechanism responsible for electron exchange in natural methanogenic communities aggregates6 and methanogenic system supplemented with electrically conductive particles such as magnetite (Fe3O4)4.
Energy efficient anaerobic fluidized membrane bioreactor (AFMBR) systems, which use granular activated carbon (GAC) as the fluidized media, are developed to treat domestic wastewaters with high energy harvest7,8,9. Effluent produced by anaerobic fluidized bed bioreactor (AFBR) was treated further by anaerobic fluidized bed ceramic membrane bioreactor (AFCMBR), termed as staged, anaerobic fluidized bed ceramic membrane bioreactor (SAF-CMBR). In these setups, GAC addition was originally conceived to serve as a mechanical scouring agent along membrane for reducing membrane fouling and a carrier for microbial attachment. Recently, we examined the microbial community in a staged anaerobic fluidized-bed ceramic membrane bioreactor (SAF-CMBR) consisting of anaerobic fluidized-bed bioreactor (AFBR) followed by anaerobic fluidized-bed ceramic membrane bioreactor (AFCMBR), fed with acetate and propionate8. This presented an ideal system for investigating syntrophic microbial interactions as these substrates are the key precursors of methanogenesis, driving IET in energy-limited methanogenic systems10,11,12. We observed the co-dominance and tight interactions between syntrophic propionate oxidizing bacteria (SPOB), Syntrophobacter and Smithella, acetoclastic methanogen Methanothrix, and exoelectrogen Geobacter on the GAC particles, suggesting that GAC serves as an electrically conductive material for promoting DIET in the SAF-CMBR8. This lends further credence to the growing literature on GAC-facilitated DIET3. Nevertheless, the metabolic interactions, particularly between Methanothrix and Geobacter, have yet to be fully understood. Such energy efficient SAF-CMBR was an ideal system to explore the underlying metabolic mechanisms and to provide their exploitation for the further improvement of the reactor design and operation.
Competition may occur between Methanothrix and Geobacter as both species utilize acetate for methanogenesis and respiration13,14. However, synergetic interaction between these two microbes is also possible. Most interestingly, while Methanothrix is unable to perform hydrogentrophic methanogenesis using CO2 due to the absence of a hydrogen uptake mechanism15, it can form syntrophic association, termed ‘electric syntrophy’, with Geobacter to achieve methane production, in a similar fashion to hydrogentrophic methanogenesis, through DIET-dependent CO2 reduction6,16. On the other hand, Kono et al. (2017) recently proposed a ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO)-mediated CO2 fixation pathway in many methanogen species including Methanothrix. It could reduce CO2 into various carbon intermediates for important metabolic pathways, such as gluconeogenesis and glycolysis, with acetate or/and hydrogen Kono, Mehrotra, Endo, Kizu, Matusda, Kimura, Mizohata, Inoue, Hasunuma, Yokota, Matsumura and Ashida17. This newly-discovered pathway, termed “reductive hexulose-phosphate (RHP) pathway”, is analogous to the Calvin–Benson cycle in plant photosynthesis, raising our hypothesis that it could be electron-driven (i.e., DIET). Additionally, this carbon fixation pathway has a potential link with methanogenesis via a formaldehyde intermediate, leading to our speculation of a third methanogenesis route in Methanothrix.
This study examines the interspecies interactions, particularly between Geobacter and Methanothrix, on GAC surfaces in the propionate- and acetate-fed SAF-CMBR. By employing a combinatorial approach of metagenomics and metatranscriptomics sequencing, this current enquiry aims to disclose potential methane formation pathways, facilitated by MIET and DIET, and their metabolic link in Methanothrix. The efficient and stable operation of methanogenic bioreactors relies heavily on syntrophic-driven IET mechanisms. A deeper understanding of such interactions is therefore critical to ultimately tie ecology to improvements in engineering operation and design.
Results and Discussion
Overview of the Metagenome Bin and Metatranscriptomes
GAC microbial communities of both AFBR and AFCMBR in the SFA-CMBR were predominated by SPOB (Syntrophobacter and Smithella), acetoclastic methanogens (Methanothrix), and the exoelectrogen Geobacter8. To further examine the metabolic interactions between these species, high quality genome bins (with >90% completeness) for these microbes were recovered through metagenomic short reads and hybrid assemblies (Tables S2). Three high-quality genome bins, AFBR_GAC_Bin72, AFCMBR_GAC_MaxBin.090 and AFBR_GAC_MaxBin.001, were phylogenetically identified to be closely related to the genomes of Syntrophobacter fumaroxidans, Methanothrix concilii and Geobacter lovleyi, respectively (Fig. S2).
The complete pathways for propionate degradation and acetate oxidation were recovered from the S. fumaroxidans MPOB (AFBR_GAC_Bin72) genome bin and G. lovleyi genome bin (AFBR_GAC_MaxBin.001), respectively. The acetoclastic methanogenesis, classical CO2 reduction and RHP pathways were also fully recovered from the M. concilii genome bin (AFCMBR_GAC_MaxBin.090) (Fig. 1; Table S2). Metatranscriptomics analysis confirmed that the genes involved in the aforementioned pathways were actively-transcribed.
Annotated pathways of acetate oxidation, acetocalstic methanogenesis, classical CO2 reduction and CO2 reduction via RHP for methane production. Expression level of involved genes were evaluated as the log2 RPKM values and represented by the bar chart. Ru5P, ribulose-5-phosphate; RuBP, ribulose-1,5-bisphosphate; 3-PGA, 3-phosphoglycerate; BPG, 1,3-diphosphoglycerate; GAP, glyceraldehyde-3-phosphate; FBP, fructose-1,6-bisphosphate; F6P, fructose-6-phosphate; Hu6P, D-arabino-3-hexulose-6-phosphate; H4MPT, tetrahydromethanopterin.
Propionate oxidation and acetoclastic methanogenesis
In our propionate and acetate-fed methanogenic system, propionate was first degraded into acetate and hydrogen, thereby acetate was adequately available as electron donor for further metabolization. Syntrophobacter degrade propionate into acetate using methylmalonyl-CoA (MMC) pathway adopted by most SPOB11. While a unique syntrophic propionate oxidizer, Smithella was transcriptionally-active and it degrades propionate into acetate via butyrate using a distinctly different pathway, of which the genes involved are still unclear so far18. On the other hand, the MMC pathway was fully reconstructed in the S. fumaroxidans MPOB genome bin, and these genes were found to be highly expressed with log2 RPKM values of 5.17–10.13 (Tables S3, S4). Therefore, such acetate-rich environment was favorable for acetate-utilizing microbes, explaining the enrichment of exoelectrogenic Geobacter and methanogenic Methanothrix. Moreover, G. lovleyi is capable to use hydrogen as an alternate electron donor and exhibits lower hydrogen consumption threshold concentration than that of methanogen14. The limited hydrogen produced from the propionate degradation pathways was too low to favor the growth of hydrogenotrophic methanogens in the system, while it could be utilized by G. lovleyi and further facilitated its dominance. This was reflected by the observation that the abundance of hydrogenotrophic methanogens was relatively low and G. lovleyi dominated in the community.
Acetate, which acts as an electron diffusive carrier of MIET, is assimilated by acetoclastic methanogens for methane generation. The acetoclastic methanogenesis pathway, which splits acetate into a methyl group and an enzyme-bound CO then further reduced to methane19, was fully reconstructed in the M. concilii genome bin (Table S3). The genes involved were also found to be highly expressed in the genome bin (log2 RPKM values of 5.45–11.96) (Table S4; Fig. 1), indicating that M. concilii was metabolically active and contributed to the methane production via its acetoclastic methanogenesis pathway.
In propionate-fed syntrophic community, the metabolic activities of SPOB and methanogens are intimately dependent on each other20. Collectively, these findings confirmed that a syntrophic interaction was present between GAC-dwelling acetoclastic methanogen M. concilii and SPOB, S. fumaroxidans MPOB and Smithella. sp., to achieve the complete bioconversion of propionate to CH4 and CO2. Given that these microbes were selectively enriched on the GAC in the SFA-CMBR8, it is most likely due to GAC facilitated MIET by enabling the microbes to grow in proximity of each other.
Acetate oxidation and DIET-dependent CO2 reduction
Pathway reconstruction in the draft genome bin related to G. lovleyi showed the capability of acetate utilization and CO2 production via the tricarboxylic acid (TCA) cycle (Fig. 1; Table S3). Gene expression patterns further confirmed that the acetate oxidation pathway was metabolically active in G. lovleyi (log2 RPKM values of 4.53–8.30) (Fig. 1; Table S4). The high abundance of gene transcripts was also observed when two Geobacter species were co-cultured together and acetate was available as an electron donor5. Both the G. lovleyi acetate oxidation and Methanothrix acetoclastic methanogenesis pathway were found to be transcriptionally-active, indicating that they were competing for acetate at the substrate-level. Concurrently and more importantly, they also formed an electric syntrophic relationship and benefited each other via the IET. It was reported that the growth of Geobacter spp. was suppressed when methanogenesis was inhibited, suggesting that Geobacter grew under syntrophic or synergetic association with methanogens16. Methanogens are possibly “electron receivers” and serve as electron sinks of the dissipated electrons from Geobacter.
Why such a substrate-competing relationship exists within a cooperative association? Is it associated with DIET-induced interactions attributed to the electrically-conductive GAC? Indeed, acetoclastic methanogenesis was not the only transcriptionally-active pathway detected in the M. concilii genome bin. The DIET-dependent CO2 reduction methanogenesis pathway was also recovered (Fig. 1; Table S3). The genes specifically associated with the CO2 reduction pathway (fwd, ftr, mch, mtd and mer) were highly expressed at levels close to the acetoclastic methanogenesis pathway (log2 RPKM values of 5.89–9.68) (Fig. 1; Table S4). Unlike hydrogenotrophic methanogens, Methanothrix is incapable of performing CO2 reduction to methane via MIET as it cannot uptake reducing equivalents in the form of hydrogen and formate5,19,21, suggesting that DIET-driven methanogenesis was prevalent within the GAC community of SFA-CMBR. This observation agrees with a finding that Geobacter species could transfer electrons to Methanothrix species to support CO2 reduction via DIET22,23. In other words, G. lovleyi and M. concilii also established a close “electric syntrophic” relationship for the generation of methane from CO2. The sources of CO2 could be extracellular (CO2 released from propionate oxidation and TCA cycle in SPOB and Geobacter, respectively) and intracellular (CO2 as a byproduct from acetoclastic methanogenesis). In M. concilii, the by-product of MIET facilitated pathway (acetoclastic methanogenesis), CO2, was further utilized in the DIET facilitated pathways (CO2 reduction). By coupling the MIET and DIET, M. concilii could utilize the metabolite, CO2, for additional energy capture. Accordingly, in this SFA-CMBR, such interspecies interactions facilitated DIET-dependent pathway and promoted the overall energy recovery in the form of methane.
CO2 reduction via the RHP pathway
Besides the classical CO2 reduction, the RHP pathway for carbon fixation is expected to be widely distributed in methanogenic archaea and the genes in such pathway are conserved in M. concilii17. Indeed, the complete RHP pathway was identified in the M. concilii genome bin (Fig. 1; Table S3). All the genes involved in the RHP pathway were at equally high expression levels as the acetoclastic and classical DIET-dependent CO2 methanogenesis pathways with log2 RPKM values ranging between 6.05 and 9.79 (Fig. 1; Table S3). This provides the first definitive proof that the entire RHP pathway is metabolically active in M. concilii. The RHP pathway, similar to the Calvin–Benson cycle, includes three phases: carbon fixation, carbon reduction, and ribulose-1,5-bisphosphate (RuBP) regeneration17. A study analyzing the RHP pathway in vivo for Methanospirillum hungatei showed that a small proportion of carbons fixed by RuBisCO were recycled for RuBP regeneration in the RHP pathway, and while a high proportion of fixed carbons were supplied to gluconeogenesis and glycolysis17. Also, it was proposed that the archaea invested a much smaller fraction of energy in the RHP pathway as compared to plant’s energy investment in the Calvin–Benson cycle17. By accomplishing this carbon fixation pathway with relatively low-energy investment, M. concilii could achieve further cell synthesis, therefore facilitating the dominance of M. concilii in the community and strengthened their overall activities. Hence, it is very likely that the RHP pathway plays an important role in M. concilii anabolism.
An important question is if the RHP pathway in M. concilii mediates methane production. The formaldehyde intermediate has been speculated to act as a metabolic link between the RHP pathway and methanogenesis in methanogens17. Accordingly, formaldehyde released from the RHP cycle can be condensed with tetrahydromethanopterin to form methyl-H4MPT, which is a key methanogenic precursor also central to both the methanogenesis and classical CO2 reduction pathways (Fig. 1). Four copies of the 5,6,7,8-tetrahydromethanopterin hydrolyase gene (fae), which perform formaldehyde condensation, were successfully recovered from the M. concilii genome bin (Fig. 1; Table S3). The high expression levels of fae (log2 RPKM values of 7.54–8.66) strongly suggest the involvement of the RHP pathway in methanogenesis. Moreover, similar to the classical DIET-dependent CO2 methanogenesis, the RHP carbon fixation pathway is also an electron-consuming process. This raises the possibility that the RHP pathway could be relying on external electrons received from G. lovleyi through DIET.
Thermodynamics estimation of the CO2 reduction pathways
The transcriptional activity of all MIET and DIET-facilitated methanogenesis pathways meant that they were all concurrently happening. Therefore, the thermodynamics of each pathway was explored. Table 1 summarizes the reactions of acetoclastic methanogenesis (Eq. 1), CO2 reduction to methane (Eq. 2), CO2 fixation via RHP pathway (Eq. 3). At biological conditions (298 K and pH 7.0), the standard Gibbs free energy changes (∆G0′) of Eq. 1–Eq. 3 were calculated (Table S5). Given that the intracellular-produced CO2 from acetoclastic methanogenesis could serve as a substrate for CO2 reduction in M. concilii, a concurrent MIET and DIET activity could result in a complete acetate reduction to methane (2 mole of methane formation per 1 mole acetate consumption). This is reflected in the summation of acetoclastic methanogenesis with classical CO2 reduction (Eq. 4) and RHP pathway (Eq. 5). As shown in Table 1, all these discussed reactions were thermodynamically favorable under standard biological conditions since all the ∆G0′ values were far below zero. Additionally, the energy released/yielded from the classical CO2 reduction (−86.95 kJ mol−1) and RHP pathway (−53.95 kJ mol−1) are significant and higher than that from acetoclastic methanogenesis, when there is an incoming electron supply for M. concilii through DIET. With facilitation of DIET, methanogens can achieve CO2 reduction and yield more energy compared to conditions without external electrons. The yielded energy in M. concilii results in more methane formation, therefore improving the overall energy recovery efficiency of the AFCMBR.
To further evaluate thermodynamic feasibility of these reactions in the AFCMBR, the transformed Gibbs free energy values (∆G′) at 298 K and pH 7 were estimated within an acetate concentration range of 0.03 mM–4 mM and a CH4/CO2 partial pressure ratio of 1–4, which mimics the actual conditions prevalent in the AFCMBR and other anaerobic digestion systems. Figure 2a displays the variation of ∆G′ for acetoclastic methanogenesis, indicating that the reaction can proceed even under very low acetate concentration. The energy gain from acetoclastic methanogenesis gently decreases as the acetate concentration decreases, while its effects on ∆G′ became more obvious at extremely low acetate concentration. In comparison, within the set range, changes of partial pressure ratio of CH4 to CO2 exerted insignificant influence on the energy gain. On the other hand, as shown in Fig. 2b, both the complete acetate reduction reactions were highly driven, and both of their energy gains (−60 to −80 and −90 to −110 kJ mol−1) were higher than that of acetoclastic methanogenesis alone without further CO2 reduction (−30 to −40 kJ mol−1). Therefore, when there are electrons available via DIET for M. concilii, the thermodynamic driving force for further CO2 reduction and/or the complete acetate reduction into methane is favorable. Notably, the energy gain from the complete acetate reaction via classical CO2 reduction is higher compared to the one via the RHP pathway, due to the differences of each metabolism involved intrinsically. Similar to the acetoclastic methanogenesis, ∆G′ of the two complete acetate reduction reactions were hardly affected by the partial pressure ratio of CH4/CO2, and their energy gains gently enhances as the acetate concentration increases. Overall, all the three reactions were thermodynamically feasible under this SFA-CMBR even at very low substrate concentrations. The thermodynamics calculation verified the feasibility of the acetoclastic methanogenesis and classical and RHP CO2 reduction pathway under such system conditions. With these two CO2 reduction pathways facilitated by DIET, the energy gain from the complete acetate reduction was higher that acetoclastic methanogenesis alone without further CO2 reduction.
Transformed Gibbs free energy values (∆G′298K) in kJ mol−1 at pH of 7 and pressure of 1 atm as a function of acetate concentration and partial pressure ratio of CH4/CO2 for (a) Eq. 1: acetoclastic methanogenesis (b) Eq. 4 (complete acetate reduction via classical CO2 reduction) and Eq. 5 (complete acetate reduction via RHP pathway). The red curve represents Eq. 4, and the red orange one represents Eq. 5.
Methods
Sample collection
The fluidized GAC samples were taken from the propionate- and acetate-fed (250 mg COD L−1) SAF-CMBR on Day 162, when the system was at steady state, for metagenomics and transcriptomics sequencing8. Genomic DNA extraction method was described in the previous study8. Shotgun metagenomic library construction and sequencing was performed on Illumina HiSeq. 4000 platform (Illumina, San Diego, CA, USA) at BGI (Shenzhen, China), generating paired-end (PE) reads with a read length of 150 base pair (bp). About 50 Gbp of metagenomic data per sample was generated. Meanwhile, 10 kb metagenomic library construction and sequencing were also performed on the PacBio Sequel Platform (Pacific Biosciences of California, Menlo Park, CA). Single-end (SE) reads with a mean insert length of 5700 base pair (bp), and 11.87 Gbp (AFBR_GAC) and 5.34 Gbp (AFCMBR_GAC) of metagenomic data were generated.
The two GAC samples were obtained from AFBR and AFCMBR for metatranscriptomics sequencing when CH4 generation reached steady phase. Experimental conditions of SAF-CMBR system are explained detail in our previous work8. The samples were preserved with RNAlater solution (Thermofisher, USA) in a volume ratio of 1:1 immediately after sample collection and frozen at −20 °C overnight. Subsequently, the samples were sent to BGI (Shenzhen, China) for total RNA extraction, metatranscriptomic library construction and metatranscriptomic sequencing on Illumina Hiseq. 4000 platform (Illumina, San Diego, CA, USA). rRNA was removed with kit after total RNA was collected from. Fragmentation buffer was added for interrupting mRNA to short fragments. Paired-end (PE) with read length of 150 bp and 15.4Gbp of metatranscriptomics data per sample was generated.
Genomic analysis
For metagenomics analysis, the raw reads were first trimmed with a minimum quality cutoff of 3, and further screened to be at least 78 bp in length, having an average quality score >30 and containing less than 3 ambiguous nucleotides (N’s) using Trimmomatic24. Followig which, digital normalization was performed to remove redundant sequences with khmer scripts (k-mer size 20). Paired-end reads were de novo assembled into long sequence contigs using St. Petersburg genome assembler (SPAdes, version 3.9.0) based on de Bruijn graph with default settings (“-k 19,33,47,61,75–careful”)25,26. Meanwhile, a parallel hybrid assembly was performed on the trimmed reads obtained from the Illumina HiSeq platform together with the trimmed reads obtained from the PacBio Sequel Platform using the same parameters. Next, MaxBin was used for binning the assembled contigs into taxonomic bins based on an Expectation-Maximization algorithm27. Then, CheckM was performed to assess the quality of draft genomes using a broader set of marker genes specific to the position of a genome within a reference genome tree and information about the collocation of these genes28. The recovered genome bins were phylogenetically identified by comparing with reference genomes using PhyloPhlAn29. Prokka (version 1.11)30 was used annotated protein coding genes. Then gene functions were further characterized and functional pathways were reconstructed with BlastKOALA31.
For metatranscriptomics analysis, the raw reads were first trimmed with a minimum quality cutoff of 3, and further screened to be at least 50 bp in length, having an average quality score >30 and containing less than 3 ambiguous nucleotides (N’s) using trimmomatic24. Assembly was performed on the paired-end trimmed reads using Trinity32. Afterward, Sequence Expression AnaLyzer (Seal) in the BBTools suite was used to map the assembled metatranscriptomic file against the recovered high-quality genome bins generated from metagenomic analysis pipeline under “ambig modes”33. Genes expression level was evaluated based on generated Reads Per Kilobase Million (RPKM) and calculated as log2 RPKM values.
References
Li, W. W. & Yu, H. Q. From wastewater to bioenergy and biochemicals via two-stage bioconversion processes: A future paradigm. Biotechnol Adv 29, 972–982 (2011).
Leng, L. et al. A review on the bioenergetics of anaerobic microbial metabolism close to the thermodynamic limits and its implications for digestion applications. Bioresource Technology 247, 1095–1106 (2018).
Liu, F. H. et al. Promoting direct interspecies electron transfer with activated carbon. Energy & Environmental Science 5, 8982–8989 (2012).
Cruz Viggi, C. et al. Magnetite particles triggering a faster and more robust syntrophic pathway of methanogenic propionate degradation. Environ Sci Technol 48, 7536–7543 (2014).
Shrestha, P. M. et al. Transcriptomic and Genetic Analysis of Direct Interspecies Electron Transfer. Appl Environ Microb 79, 2397–2404 (2013).
Morita, M. et al. Potential for Direct Interspecies Electron Transfer in Methanogenic Wastewater Digester Aggregates. Mbio 2 (2011).
McCarty, P. L., Bae, J. & Kim, J. Domestic Wastewater Treatment as a Net Energy Producer-Can This be Achieved? Environmental Science & Technology 45, 7100–7106 (2011).
Aslam, M., Yang, P., Lee, P. H. & Kim, J. Novel staged anaerobic fluidized bed ceramic membrane bioreactor: Energy reduction, fouling control and microbial characterization. J Membrane Sci 553, 200–208 (2018).
Kim, J. et al. Anaerobic Fluidized Bed Membrane Bioreactor for Wastewater Treatment. Environmental Science & Technology 45, 576–581 (2011).
de Bok, F. A. M., Plugge, C. M. & Stams, A. J. M. Interspecies electron transfer in methanogenic propionate degrading consortia. Water Research 38, 1368–1375 (2004).
Müller, N., Worm, P., Schink, B., Stams, A. J. & Plugge, C. M. Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanisms. Environmental microbiology reports 2, 489–499 (2010).
McCarty, P. L. The development of anaerobic treatment and its future. Water Science and Technology 44, 149–156 (2001).
Patel, G. B. & Sprott, G. D. Methanosaeta-Concilii Gen-Nov, Sp-Nov (Methanothrix-Concilii) and Methanosaeta-Thermoacetophila Nom-Rev, Comb-Nov. Int J Syst Bacteriol 40, 79–82 (1990).
Sung, Y. et al. Geobacter lovleyi sp nov strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl Environ Microb 72, 2775–2782 (2006).
Berg, I. A. et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol 8, 447–460 (2010).
Kato, S., Hashimoto, K. & Watanabe, K. Methanogenesis facilitated by electric syntrophy via (semi)conductive iron-oxide minerals. Environ Microbiol 14, 1646–1654 (2012).
Kono, T. et al. A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nat Commun 8 (2017).
de Bok, F. A., Stams, A. J., Dijkema, C. & Boone, D. R. Pathway of Propionate Oxidation by a Syntrophic Culture of Smithella propionica andMethanospirillum hungatei. Applied and environmental microbiology 67, 1800–1804 (2001).
Smith, K. S. & Ingram-Smith, C. Methanosaeta, the forgotten methanogen? Trends Microbiol 15, 150–155 (2007).
Kato, S. & Watanabe, K. Ecological and Evolutionary Interactions in Syntrophic Methanogenic Consortia. Microbes Environ 25, 145–151 (2010).
Kosaka, T. et al. Reconstruction and regulation of the central catabolic pathway in the thermophilic propionate-oxidizing syntroph Pelotomaculum thermopropionicum. J Bacteriol 188, 202–210 (2006).
Holmes, D. E. et al. Metatranscriptomic evidence for direct interspecies electron transfer between geobacter and methanothrix species in methanogenic rice paddy soils. Applied and Environmental Microbiology 83, e00223–00217 (2017).
Zhang, S. et al. Enhancement of methanogenesis via direct interspecies electron transfer between Geobacteraceae and Methanosaetaceae conducted by granular activated carbon. Bioresource technology 245, 132–137 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol 19, 455–477 (2012).
Nurk, S. et al. In Research in Computational Molecular Biology: 17th Annual International Conference, RECOMB 2013, Beijing, China, April 7–10, 2013. Proceedings. (eds M. Deng, R. Jiang, F. Sun & X. Zhang) 158–170 (Springer Berlin Heidelberg, Berlin, Heidelberg; 2013).
Wu, Y.W., Tang, Y.H., Tringe, S.G., Simmons, B.A. & Singer, S.W. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2 (2014).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25, 1043–1055 (2015).
Segata, N., Bornigen, D., Morgan, X.C. & Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4 (2013).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Kanehisa, M., Sato, Y. & Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J Mol Biol 428, 726–731 (2016).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–U130 (2011).
Bushnell, B. BBTools software package. http://sourceforge.net/projects/bbmap (2014).
Acknowledgements
The authors wish to acknowledge the Research Grants Council (RGC) General Research Fund (15273316), Collaborative Research Fund (C7044-14G) and Theme-based Fund (T21-711/16-R) for providing financial support. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science & ICT (2017RA1A2B4007804). The first author gratefully acknowledges RGC for the support of her higher degree under the Hong Kong Ph.D. Fellowship (PF13-12713).
Author information
Authors and Affiliations
Contributions
P.Y. and G.Y.T.: Data analysis, wrote the manuscript. M.A.: reactors operation. P.Y., G.Y.T., M.A., J.K. and P.H.L.: reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, P., Tan, GY.A., Aslam, M. et al. Metatranscriptomic evidence for classical and RuBisCO-mediated CO2 reduction to methane facilitated by direct interspecies electron transfer in a methanogenic system. Sci Rep 9, 4116 (2019). https://doi.org/10.1038/s41598-019-40830-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-40830-0
This article is cited by
-
Light-independent anaerobic microbial oxidation of manganese driven by an electrosyntrophic coculture
The ISME Journal (2023)
-
Adaptation of a microbial community to demand-oriented biological methanation
Biotechnology for Biofuels and Bioproducts (2022)
-
Wind speed pattern data and wind energy potential in Pakistan: current status, challenging platforms and innovative prospects
Environmental Science and Pollution Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.