Abstract
Global warming has profound effects on plant growth and fitness. Plants have evolved sophisticated epigenetic machinery to respond quickly to heat, and exhibit transgenerational memory of the heat-induced release of post-transcriptional gene silencing (PTGS). However, how thermomemory is transmitted to progeny and the physiological relevance are elusive. Here we show that heat-induced HEAT SHOCK TRANSCRIPTION FACTOR A2 (HSFA2) directly activates the H3K27me3 demethylase RELATIVE OF EARLY FLOWERING 6 (REF6), which in turn derepresses HSFA2. REF6 and HSFA2 establish a heritable feedback loop, and activate an E3 ubiquitin ligase, SUPPRESSOR OF GENE SILENCING 3 (SGS3)-INTERACTING PROTEIN 1 (SGIP1). SGIP1-mediated SGS3 degradation leads to inhibited biosynthesis of trans-acting siRNA (tasiRNA). The REF6-HSFA2 loop and reduced tasiRNA converge to release HEAT-INDUCED TAS1 TARGET 5 (HTT5), which drives early flowering but attenuates immunity. Thus, heat induces transmitted phenotypes via a coordinated epigenetic network involving histone demethylases, transcription factors, and tasiRNAs, ensuring reproductive success and transgenerational stress adaptation.
Similar content being viewed by others
Introduction
Increasing global temperatures have diverse and profound effects on plant growth, development and reproduction, and greatly threaten global crop yields.1,2 Plants have evolved sophisticated epigenetic machinery to respond quickly to heat.3 Heat exposure activates various transgenes and repetitive elements that are normally silenced via transcriptional gene silencing (TGS).4,5,6,7 Heat stress can also release post-transcriptional gene silencing (PTGS) by inhibiting siRNA biogenesis.8,9,10 Importantly, heat stress decreases the abundance of trans-acting siRNAs (tasiRNAs) derived from eight Arabidopsis Trans-acting siRNA (TAS) loci (TAS1a-c, TAS2, TAS3a-c and TAS4), triggering the upregulation of HEAT-INDUCED TAS1 TARGET (HTT) genes, which mediate thermotolerance by acting as cofactors of HEAT SHOCK PROTEIN 70–14 (Hsp70–14).8 It has been widely recognized that DNA methylation is not involved in the heat-induced release of gene silencing.3
Heat responses are usually transient and reset quickly to the normal conditions, although there are some somatic stress memory with the duration of several days.11 Two ATP-dependent chromatin remodeling factors, Decrease in DNA methylation1 (DDM1) and Morpheus’Molecule 1 (MOM1), have been shown to act in inhibiting the transgenerational transmission of heat-induced epigenetic states.12 Moreover, the transgenerational retrotransposition of ONSEN is prevented by the small interfering RNAs (siRNAs) pathway.6 However, certain plant responses to extreme or prolonged heat stress have been shown to exhibit transgenerational memory as they can be detectable in one or two subsequent stress-free generations.7,10,13 For instance, the immediate progeny of extreme heat shock (50 °C, 3 h/day for 5 days)-stressed Arabidopsis plants tend to bolt earlier.13 Heat stress (42 °C for 48 h)-mediated release of a reporter gene silencing can be transmitted to the non-stressed progeny, which was restricted to a small number of cells and limited to only two non-stressed progeny generations.7 Overall, the precise molecular mechanisms underlying the transgenerational memory of heat stress in plants remain poorly understood and disputed.
We previously reported that prolonged high temperature (30 °C for 13 days) led to deterministic suppression and transgenerational inhibition of PTGS and tasiRNA biogenesis in Arabidopsis.10 Our data suggested that heat stress reduces the steady-state level but not the transcription level of SUPPRESSOR OF GENE SILENCING 3 (SGS3), a plant-specific RNA binding protein critical for siRNA biogenesis.14 However, how the memory of heat stress is established and transmitted to progeny and whether other phenotypes are affected remained largely unknown.
Here, we discover that HEAT SHOCK TRANSCRIPTION FACTOR A2 (HSFA2) and H3K27me3 demethylase RELATIVE OF EARLY FLOWERING 6 (REF6) form a positive feedback loop to transmit long-term epigenetic memory of heat by maintaining the transgenerational “ON state” of HSFA2, which activates an F-Box E3 ubiquitin ligase gene, SGS3-INTERACTING PROTEIN 1 (SGIP1), to execute the transgenerational degradation of SGS3. More importantly, the thermomemory machinery promotes plant reproduction and fitness by accelerating flowering and attenuating disease resistance through the tasiRNA target HTT5.
Results
Transgenerational memory of early flowering and attenuated immunity via suppression of tasiRNAs
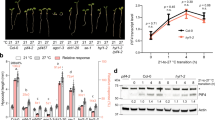
Extreme heat (50 °C)-stressed plants show accelerated flowering, which is also observed in the immediate unstressed progeny, however, the underlying mechanism remains unknown.13 To investigate whether the prolonged high temperature (30 °C) has similar effect in the progeny, we generated prolonged heat-stressed seedlings (Supplementary information, Fig. S1a) and analyzed the flowering time in their progeny. Interestingly, we found that parental (first generation) Col plants grown at prolonged high temperature bolted earlier than those grown at 22 °C, and that most of their unstressed second generation progeny also blotted earlier (Fig. 1a). Although there was no statistical significance, part of third generation progeny still tended to bolt earlier (Fig. 1a), similar to the heat-induced PTGS release which was maintained for at least three generations with rapidly declining strength.10 High temperatures accelerate flowering via induction of PHYTOCHROME INTERACTING FACTOR4 (PIF4), which activates florigen FLOWERING LOCUS T (FT).15 Indeed, FT was upregulated in heat-stressed wild-type Col and unstressed progeny both before and after blotting (Fig. 1b; Supplementary information, Fig. S1b). In contrast, although high temperatures induced PIF4 expression, PIF4 upregulation was not detected in unstressed progeny, indicating that factors other than PIF4 are involved in the transgenerational thermomemory (Fig. 1b; Supplementary information, Fig. S1b, c).
Heat-induced transgenerational degradation of SGS3 accelerates flowering but attenuates immunity. a Four-week-old 22 °C-grown Col, heat-stressed (first) and unstressed second and third generation plants and box plots of flowering times of these four lines. Flowering time was assessed by counting total leaf numbers in bolting plants (n ≥ 15 for each line). Bars represent means ± SD. b PIF4 and FT transcript levels as normalized to the ACTIN2 signals. The average values (±SD, n = 3) were shown. Samples were collected from 17-day-old seedlings. Asterisks indicate significant difference (Student’s t-test; *P < 0.05, **P < 0.01, ***P < 0.001) (a, b). c Disease symptoms at 4 dpi with Pst DC3000 (avrRpt2). Scale bar, 1 cm (a, c). d Bacterial growth in plants in c was measured at 0 and 4 dpi. Bars represent means ± SD (n = 3). Two-way ANOVA with Tukey’s HSD post hoc test (significance set at P < 0.05) was performed. Different lowercase letters indicate significant differences. e SGS3 levels were immunodetected, with relative levels shown under lanes. Samples were collected from 24-day-old 22 °C-grown Col, heat-stressed (first) and early flowering, unstressed second and third generation plants. Rubisco served as a loading control
High temperatures are known to diminish the resistance to bacterial pathogens through inhibiting the salicylic acid (SA) defense pathway or upregulating PIF4.16,17 Interestingly, we observed transgenerational memory of heat-induced enhanced susceptibility to avirulent Pseudomonas syringae (P. syringae) Pst DC3000 (avrRpt2) (Fig. 1c, d). Thus, heat stress induces a memory of attenuated immunity, along with early flowering and PTGS release. Since PIF4 is not transgenerationally upregulated (Fig. 1b; Supplementary information, Fig. S1b, c), we then investigated the SA pathway. Inoculated with Pst DC3000 (avrRpt2), heat-stressed plants showed much lower inductions of the SA biosynthesis gene ISOCHORISMATE SYNTHASE 1 (ICS1), the SA signaling marker PATHOGENESIS-RELATED GENE 1 (PR1), and SA, but their levels were partially restored in unstressed progeny (Supplementary information, Fig. S2a–c).
Since heat stress reduces the levels of SGS3 (Fig. 1e) and tasiRNAs,10 we hypothesized that reduced SGS3 and tasiRNA levels might result in the transgenerational memory of early flowering and attenuated immunity, similar to PTGS release.10 We observed reduced levels of endogenous SGS3, siR255 and siR1511 (two widely studied tasiRNAs that derive from TAS1 and TAS2, respectively)18 in heat-stressed first generation Col plants, and in early flowering unstressed second and third generation plants (Fig. 1e; Supplementary information, Fig. S3a). We also found that the loss-of-function sgs3–12 mutant19 and MIM480(+) plants, which have reduced levels of TAS1-siRNAs,8 also bolted earlier when grown at 22 °C and were not affected by heat, associated with upregulated FT (Supplementary information, Fig. S3b, c). These results suggest that the heat-induced decrease in tasiRNAs is involved in the thermomemory of early flowering.
We next examined whether the reduced SGS3 and tasiRNA levels also contribute to the transgenerational memory of attenuated immunity. Indeed, sgs3–12 mutant and MIM480(+) plants were more susceptible to Pst DC3000 (avrRpt2) relative to wild-type plants (Supplementary information, Fig. S3d), with only slightly reduced induction of ICS1 and SA levels upon pathogen infection (Supplementary information, Fig. S3e, f). These results suggest that depletions of SGS3 and tasiRNAs compromise immunity, which may be only partially dependent on the SA pathway. Thus, the heat-induced memory of attenuated immunity is likely caused by defects in multiple defense pathways. Based on these data, we propose that thermomemory affects the fitness of stressed plants and their unstressed progeny by accelerating reproductive development associated with attenuated immunity through certain tasiRNA target(s), consistent with the trade-off between growth and defense.20,21
SGIP1 targets SGS3 for degradation
Our findings so far suggest that heat stress triggers transgenerational inhibition of SGS3 (Fig. 1e). In a cell-free degradation assay, we observed faster degradation of SGS3 isolated from heat-stressed seedlings, and delayed SGS3 degradation with treatment of the proteasome inhibitor MG132 (Supplementary information, Fig. S4a). The heat-enhanced degradation of SGS3 was further confirmed in Col seedlings treated with cycloheximide (CHX), which can block new protein synthesis, and MG132 treatment inhibited SGS3 degradation both at 22 °C and 30 °C (Supplementary information, Fig. S4b). These results suggest that a heat-upregulated E3 ligase targets SGS3 for degradation. Therefore, we analyzed published data22,23 and pursued 46 putative heat-responsive E3 ligases, which could be induced upon heat treatment (Supplementary information, Table S1).
We screened the homozygous mutants of 46 putative heat-responsive E3 ligases at 22 °C and 30 °C and identified that a knockdown mutant of AT3G47020 showed impaired heat-induced decrease in SGS3 abundance (Fig. 2a; Supplementary information, Fig. S4c), and delayed SGS3 degradation compared to Col in vivo and in vitro (Supplementary information, Fig. S4d, e), leading us to investigate the possibility that this E3 ligase interacts with SGS3 to trigger its degradation. We found that AT3G47020 and SGS3 co-localized in the cytoplasmic granules (Supplementary information, Fig. S5a), and interacted in the yeast two-hybrid assay and the split luciferase complementation assay (Supplementary information, Fig. S5b, c). Importantly, SGS3 co-immunoprecipitated with FLAG-AT3G47020 in planta (Fig. 2b). These results suggest that AT3G47020 physically interacts with, and may directly regulates, SGS3. Thus, we refer to this putative E3 as SGS3-INTERACTING PROTEIN 1 (SGIP1) and the knockdown mutant of AT3G47020 as sgip1.
SGIP1 mediates thermomemory degradation of SGS3 and tasiRNA suppression. a Immunodetection of SGS3 abundance in 24-day-old plants. Coomassie blue staining of Rubisco served as a loading control. b Co-IP assays of SGIP1 with SGS3 in N. benthamiana leaves. Proteins were immunoprecipitated with an anti-FLAG antibody, and detected by western blot using the anti-SGS3 and anti-FLAG antibodies. c In vitro ubiquitination of SGS3 by SGIP1. Polyubiquitination of SGS3 by SGIP1 was detected by immunoblotting using the anti-SGS3 antibody. The mutated SGIP1 protein (SGIP1△Fbox) served as a negative control. d SGIP1 promotes SGS3 polyubiquitination and degradation in vivo. Co-expression of SGIP1 and SGS3 was performed in N. benthamiana leaves. Protein accumulation of SGS3 and FLAG-SGIP1 was immunodetected with the anti-SGS3 and anti-FLAG antibody, respectively. GFP was co-expressed as an internal control. e Expression levels of SGIP1 as normalized to the ACTIN2 signals. Error bars indicate the SD (n = 3). Asterisks indicate significant difference between lines and 22 °C-grown Col (Student’s t-test; *P < 0.05, **P < 0.01). f Immunodetection of SGIP1 levels with an anti-SGIP1 antibody. ACTIN served as a loading control (d, f). g Knockdown of SGIP1 maintained the production of siR255 and siR1511 at 30 °C. U6 was used as a loading control. The intensity of the blots was quantified (a, f, g)
To test whether SGIP1 has E3 ligase activity, we expressed and purified an MBP-SGIP1 fusion protein from E. coli. MBP-SGIP1 can polyubiquitinate SGS3 immunopurified from N. benthamiana (Fig. 2c). Further, we detected polyubiquitinated SGS3 in N. benthamiana leaves co-expressing SGS3 with FLAG-SGIP1 (Fig. 2d). These results reveal that SGIP1 targets SGS3 for ubiquitination.
SGIP1 expression was upregulated in heat-stressed plants and unstressed progeny (Fig. 2e, f). To determine if SGIP1 upregulation is sufficient to induce SGS3 degradation, we engineered Col plants overexpressing SGIP1 (SGIP1-OE) (Fig. 2f; Supplementary information, Fig. S4c), which displayed elongated, downwardly curled leaves, bolted earlier and showed enhanced disease susceptibility, similar to sgs3–12 mutant plants (Supplementary information, Figs. S3b, d and S6a-c). Importantly, SGIP1-OE plants had substantially reduced levels of SGS3, siR255 and siR1511 (Fig. 2a; Supplementary information, Fig. S6d). Further, AT1G62590, AUXIN RESPONSE FACTOR4 (ARF4) and MYB DOMAIN PROTEIN 75 (MYB75), which are targeted by TAS2, TAS3 and TAS4-derived tasiRNAs, respectively,24,25 were upregulated in SGIP1-OE plants, as in sgs3–12 plants (Supplementary information, Fig. S6e). In contrast, similar to p35S::SGS3-GFP plants reported previously,10 heat-stressed sgip1 plants neither bolted earlier (Supplementary information, Fig. S6a, b), nor showed reduced siR255 and siR1511 levels (Fig. 2g), but showed diminished upregulation of the tasiRNA targets (Supplementary information, Fig. S6e). Thus, SGIP1 is a heat-induced negative regulator of SGS3 and, in turn tasiRNAs. Collectively, the results reveal that SGIP1 targets SGS3 for ubiquitin-dependent degradation, and that the heat-induced activation of SGIP1 is inherited in progeny. The SGIP1-OE plants blotted earlier, while the flowering time of sgip1 mutant plants showed no thermomemory, suggesting that SGIP1 is important for transgenerational thermomemory.
H3K27me3 demethylation confers transgenerational upregulation of HSFA2 and its target SGIP1
We noticed that the SGIP1 promoter contains four HSF-targeting heat shock elements (HSE).26 We hypothesized that HSFs regulate SGIP1. Indeed, HSFA2, HSFA3 and HSFA7a were obviously upregulated at 30 °C (Supplementary information, Fig. S7a), and HSFA2, a major heat stress transcription factor,3,26 remained upregulated in the unstressed progeny (Fig. 3a, b; Supplementary information, Fig. S7b). Further, we detected direct binding of HSFA2 to the SGIP1 promoter HSEs in vitro (Fig. 3c; Supplementary information, Fig. S7c). In addition, ChIP-qPCR of hsfa2 knockout plants expressing a pHSFA2::HSFA2-YFP fusion revealed that HSFA2-YFP bound to SGIP1 P1-P3 at 22 °C, and this binding increased and extended to P4 under heat stress (Fig. 3c). The expression of SGIP1 was dramatically decreased in hsfa2 plants (Fig. 3d), whereas transgenic plants overexpressing HSFA2-MYC displayed early flowering as well as elevated SGIP1 and a concomitant decrease in SGS3 (Fig. 3e; Supplementary information, Fig. S7d, e). These data suggest that upregulation of HSFA2 in heat-stressed plants and unstressed progeny leads to direct activation of SGIP1.
HSFA2 activates SGIP1 transgenerationally through binding to its promoter. a, b HSFA2 remained upregulated in the progeny of heat-stressed plants at both transcript (a) and protein (b) levels. c HSFA2 binds to the SGIP1 promoter HSEs in vitro and in vivo. Left: EMSA shows the direct binding of His-HSFA2 to the HSE-P4 of SGIP1. The arrow indicates the shifted bands. Excess unlabeled probe (500 ng and 1000 ng) outcompeted the labeled probe (right lanes). Sequence of the probe is shown and the motif is highlighted in red. A mutant HSE and a downstream sequence SGIP1-NC (negative control) were used as negative controls. Right: ChIP-qPCR validation of HSFA2 occupancy at the HSE regions of the SGIP1 promoter. Data are shown as relative fold enrichments over the background (Col). The SGIP1-NC locus was used as the negative-control locus. The locations of 4 HSEs are indicated by triangles above the gene model. The regions validated by ChIP-qPCR were marked by a bar below the gene model. d SGIP1 transcript levels in Col and hsfa2 plants grown at 22 °C and 30 °C. Gene expression was shown as mean ± SD (n = 3). ACTIN2 was analyzed as an internal control (a, d). Lowercase letters indicate significant differences, as determined by the post hoc Tukey’s HSD test (a, c, d). e The abundance of SGIP1 protein increased in three transgenic lines overexpressing HSFA2-MYC, as immunodetected with anti-SGIP1 and anti-MYC antibodies. ACTIN served as a loading control (b, e) and the signals were quantified (b)
We previously found that changes in DNA methylation did not correlate with the heat-induced transgenerational release of PTGS.10 To investigate whether other epigenetic mechanisms contribute to the regulation of HSFA2, we evaluated various histone modifications, and found that unstressed progeny inherited a heat-induced depletion of H3K27me3 within the coding regions of HSFA2 (Fig. 4a), which may contribute to the upregulation of HSFA2 in heat-stressed plants and unstressed progeny.
H3K27me3 demethylation controls transgenerational upregulation of HSFA2. a H3K27me3 levels detected by ChIP-qPCR at the HSFA2 loci. The gene model was shown and the analyzed regions (P1 and P2) were indicated. One intergenic region (NC3) without H3K27me3 mark28 was used as negative background. b EMSA showed HSFA2 probe binding to GST-REF6C containing the C2H2-ZnF cluster. The mutant probe abolished the binding of GST-REF6C. Excess unlabeled probe outcompeted the labeled probe (right panel). The motif is highlighted in red in the probe sequence, with mutated bases shown in lowercase. The region validated by ChIP-qPCR in c was marked by a bar. c ChIP-qPCR validation of REF6 binding at HSFA2 using the REF6-GFP plants. Col was used as the negative control. The TA3 locus was used as the negative-control locus. d Transcript levels of REF6 and BRM, shown as mean ± SD (n = 3). e ChIP-qPCR validation of BRM binding at HSFA2 using the BRM-GFP plants. Col was used as the negative control. f H3K27me3 levels of HSFA2 in the HSFA2Δ and ref6–1 plants. The data were normalized to the corresponding input fraction (a, c, e, f). g HSFA2 transcript levels. Data were shown as means ± SD from three replicates (a, c–g). h Flowering times of indicated lines were assessed (n ≥ 15 for each line). Lowercase letters indicate statistical significance based on one-way (a, c–f) or two-way (g, h) ANOVA with Tukey’s HSD post hoc analysis (P < 0.05)
REF6 recognizes a CTCTGYTY (Y = T or C) motif to erase H3K27me3.27,28 We identified two of these motifs within HSFA2 intron 1. We found that REF6 bind to the HSFA2 region in vitro (Fig. 4b), and validated the binding of REF6 in vivo by ChIP-qPCR (Fig. 4c). REF6 was upregulated by heat and its activation was maintained transgenerationally (Fig. 4d). REF6 recruits the chromatin remodeler BRAHMA (BRM) to activate gene expression.27 Consistent with this, BRM showed heat-induced transgenerational upregulation and the binding of BRM to the HSFA2 region was enhanced by heat (Fig. 4d, e). To evaluate the role of REF6/BRM binding in the transgenerational upregulation of HSFA2, we used the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated 9 (Cas9) technology to delete a 104-bp fragment containing the two REF6 binding motifs from HSFA2 intron 1 (Supplementary information, Fig. S8a). Similar to ref6–1 mutants, the HSFA2Δ plants displayed elevated H3K27me3 levels and reduced expression of HSFA2 (Fig. 4f, g), suggesting that these motifs directly recruit REF6/BRM to regulate H3K27me3 levels and HSFA2 expression. The flowering time of ref6–1 mutant plants showed no thermo-responsiveness or thermomemory (Fig. 4h), suggesting that the thermomemory of early flowering is dependent on the H3K27me3 epigenetic regulation. Intriguingly, hsfa2 and HSFA2Δ plants also trended early flowering, but they likely lost the thermomemory on early flowering (Supplementary information, Fig. S8b). Together, these data indicate that heat exposure reduces H3K27me3 at HSFA2 via REF6/BRM, which is inherited in unstressed progeny.
A heritable REF6-HSFA2 feedback loop enables cooperative gene activation
Interestingly, REF6 and BRM also contains HSEs within their promoter regions (Fig. 5a; Supplementary information, Fig. S9a). We postulated that upregulation of HSFA2 would thus in turn activate REF6 and BRM. Indeed, HSFA2 directly binds to one HSE within the REF6 promoter, and to all three HSEs within the BRM promoter in vitro (Fig. 5a; Supplementary information, Fig. S9a). Further, HSFA2-YFP was enriched at the HSEs within the REF6 and BRM promoters in heat-stressed plants (Fig. 5b; Supplementary information, Fig. S9b). The hsfa2 knockout plants displayed reduced expression of these genes (Fig. 5c; Supplementary information, Fig. S9c). Together, these data strongly suggest a positive feedback loop, in which heat-induced activation of HSFA2 triggers upregulation of REF6 and BRM, which in turn upregulates HSFA2. Our data also suggest that progeny inherit upregulation of the REF6/BRM-HSFA2 feedback loop as a determinant upstream node of the thermomemory.
REF6 and HSFA2 form a regulatory loop that is maternally transmitted. a EMSA shows that His-HSFA2 protein but not His by itself specifically binds to the first HSE (nTTCnnGAAn motif) of REF6 promoter in vitro. Arrows indicated the shifted bands. Excess unlabeled probe could outcompete the labeled probe (middle). A mutant HSE and a REF6-NC sequence were used as negative controls (right).The motif is highlighted in red in the probe. The region validated by ChIP-qPCR in b was marked by a bar. b ChIP-qPCR validation of HSFA2 occupancy at the REF6 promoter region. The data were normalized to the corresponding input fraction. The REF6-NC locus was used as the negative-control locus. c REF6 transcript level in 24-day-old Col and hsfa2 grown at 22 °C and 30 °C. d The progeny derived from crosses (F1) of Col 30 °C♀×Col 22 °C♂ but not Col 22 °C♀×Col 30 °C♂flowered earlier. Flowering time of different lines was assessed by counting leaf numbers in bolting plants (n ≥ 15 for each line). e Lower H3K27me3 levels of HSFA2 persisted in the progeny derived from Col 30 °C♀×Col 22 °C♂but not Col 22 °C♀×Col 30 °C♂. Data were shown as means ± SD from three replicates (b, c, e). Lowercase letters indicate statistical significance based on one-way (b, d, e) or two-way (c) ANOVA with Tukey’s HSD post hoc analysis (P < 0.05)
To determine the parental contribution in transgenerational H3K27me3 demethylation, we made reciprocal crosses between 22 °C-grown Col and 30 °C-grown Col, and analyzed the flowering time of their progeny. The progeny derived from Col 30 °C♀×Col 22 °C♂flowered as early as the progeny of 30 °C-grown Col and accumulated higher levels of FT as well as HSFA2, SGIP1 and BRM transcripts, while the progeny of Col 22 °C♀×Col 30 °C♂flowered at similar time to 22 °C-grown Col (Fig. 5d; Supplementary information, Fig. S9d). The lower H3K27me3 levels of HSFA2 were also found in the progeny of Col 30 °C♀×Col 22 °C♂ (Fig. 5e). The results demonstrate that H3K27me3 demethylation is most likely transmitted maternally.
To further investigate the role of H3K27me3 demethylation in thermomemory, we evaluated the H3K27me3 levels and expression of a set of REF6 targets including HSFA3 and HSFA7a.27,28 These REF6 targets displayed decreased H3K27me3 levels, but their expression levels were not upregulated transgenerationally (Supplementary information, Figs. S7b and 10a-c). We hypothesized that H3K27me3 demethylation alone may be insufficient to upregulate gene expression transgenerationally. We then investigated the expression of other reported HSFA2 target genes26 and observed transgenerational upregulation of 11/26 HSFA2 target genes (Supplementary information, Fig. S10d, e). Interestingly, similar to HSFA2 itself, all of these thermomemory genes, except Stress-inducible protein 1(Sti1), showed heat-induced depletion of H3K27me3 that was generally transmitted to the progeny (Supplementary information, Fig. S10f). The biological importance of these thermomemory genes is worth further investigation. These data imply that heritable REF6/BRM module-mediated H3K27me3 demethylation and HSFA2 activation cooperate to maintain the transgenerational “ON state” of thermomemory genes including HSFA2.
HTT5 coordinates thermomemory phenotypes downstream of reduced tasiRNAs and the REF6-HSFA2 loop
To further uncover the tasiRNA target(s) that functions in the heat-induced memory of early flowering and immunity inhibition as described above, we analyzed the H3K27me3 status and expression of tasiRNA target genes. We found that the siR255 target HTT5 is a co-target of HSFA2 and REF6/BRM, and its expression is regulated by H3K27me3 (Fig. 6a, b; Supplementary information, Fig. S11a-d). Further, heat-induced upregulation of HTT5 was inherited by progeny (Fig. 6c). Importantly, the htt5 mutant plants displayed neither heat-sensitive early flowering nor enhanced disease susceptibility to Pst DC3000 (avrRpt2) (Fig. 6d–f; Supplementary information, Fig. S12a, b). Further, we generated transgenic plants overexpressing a siR255-resistant version of HTT5 (rHTT5-MYC) and found that they flowered earlier with upregulated FT expression (Fig. 6d; Supplementary information, Fig. S12c, d). Compared to Col, the rHTT5-MYC lines were also more susceptible to Pst DC3000 (avrRpt2) (Fig. 6e, f). These results reveal that HTT5, the tasiRNA target, plays a dual role in growth and immunity: positively regulating reproductive development but negatively regulating immunity (Fig. 7). How HTT5 coordinates the heat-induced changes in development and immunity remains to be determined.
HTT5 coordinates thermomemory phenotypes via reduced tasiRNAs and the REF6-HSFA2 loop. a ChIP-qPCR validation of HSFA2 (left), REF6 (middle) and BRM (right) occupancy at the HTT5 loci. The analyzed regions were marked by bars. b HTT5 transcript levels in 24-day-old Col, hsfa2, ref6–1, brm-1 and ref6–1brm-1 grown at 22 °C and 30 °C. c Heat-induced upregulation of HTT5 could be transmitted to progeny, as detected by qRT-PCR. d Flowering times of indicated lines were assessed (n ≥ 15 for each line). e Disease symptoms at 4 dpi with Pst DC3000 (avrRpt2). Scale bar, 1 cm. f Bacterial growth was measured. Data are shown as means ± SD from three replicates (a, b, c, f). Lowercase letters indicate statistical significance based on one-way (a, c, d) or two-way (b, f) ANOVA with Tukey’s HSD post hoc analysis (P < 0.05)
A proposed model depicting transgenerational thermomemory that is mediated by the H3K27me3 demethylation-HSFA2 regulatory loop in Arabidopsis. Long-term heat exposure activates plant heat sensor(s) that upregulates HSFA2, which targets and activates H3K27me3 demethylase REF6 and the chromatin remodeler BRM. REF6/BRM in turn triggers H3K27me3 demethylation to activate HSFA2. Thus, REF6 and HSFA2 form a heritable transcriptional feedback loop in heat responses and memory. This regulatory loop activates the E3 ligase, SGIP1, to trigger the transgenerational degradation of SGS3, leading to the suppression of tasiRNA biosynthesis. The REF6-HSFA2 loop and reduced tasiRNA levels converge to activate HTT5, which coordinates early flowering with decreased disease resistance in heat-stressed plants and progeny
Discussion
Here, we reveal that reduced H3K27me3 and tasiRNA biogenesis are necessary and sufficient to induce several transgenerational responses to heat stress, including early flowering, attenuated immunity, and PTGS release. We discovered an E3 ligase, SGIP1, which is upregulated by mild heat and mediates ubiquitination and degradation of SGS3 transgenerationally, and controls tasiRNA biogenesis. HSFA2 activates the expression of SGIP1, and HSFA2 is activated by heat-induced demethylation of H3K27me3. Importantly, HSFA2 upregulates the expression of H3K27me3 demethylase REF6, establishing a positive feedback loop between H3K27me3 demethylation and HSFA2 upregulation that is critical to the thermomemory establishment and transmission to progeny, which further supports the transgenerational inheritance of gene activation at several target genes. In particular, heat exposure upregulates the tasiRNA target HTT5 to promote early flowering but inhibit immunity, and the thermomemory phenotypes are transmitted to unstressed progeny (Fig. 7). Importantly, we reveal the complex epigenetic mechanisms that control transgenerational degradation of SGS3 and ensure the thermomemory phenotypes via the upregulation of the tasiRNA target HTT5. These findings provide a paradigm for understanding how environmental exposure can influence the evolutionary fitness of progeny in plants.
Whether and how histone modifications function in stress memory is a mystery. Recently, histone modifications were revealed as a conserved mechanism underlying long-term transmission of the silenced state, including H3K27me3 in Drosophila and H3K9 methylation in Saccharomyces.29,30,31,32 Interestingly, in Caenorhabditis elegans, heat stress-induced H3K9me3 demethylation and gene activation can persist for at least 14 generations.33 Most recently, the reduction of H3K9me3 and H3K27me3 was found associated with the transgenerational derepression of a silenced transgene in C. elegans exposed to the chemical Bisphenol A, dependent on demethylases.34 In Arabidopsis, prolonged exposure to cold results in the mitotically heritable but meiotically intransmissible silencing of FLC through the maintenance of H3K27me3.35 Cold-triggered and H3K27me3-mediated silencing is reset during each generation.36 Therefore, the meiotic ‘memory of heat’ observed in our study is distinct from the mitotic ‘memory of cold’. We found that H3K27me3 demethylation transmits thermomemory maternally. Similar maternal contribution was also observed in embryogenesis,37 and seed dormancy entrained by maternal temperature history in Arabidopsis.38,39 Interestingly, H3K27me3-maintained transgenerational silencing is also maternally inherited in Drosophila.40 Thus, H3K27me3 or H3K9me2/3 may be conserved heritable epigenetic marks that convey stress memories and adaptation in plants and animals.
Notably, we revealed that thermomemory has a biologically important impact on plant adaptation, given the early flowering of progeny grown at the normal temperature. Interestingly, heat stress could increase seed production in a later generation.41 These findings suggest that heat exposure promotes species survival by enhancing reproduction. However, we found that unstressed progeny displayed attenuated disease resistance, suggesting that heat increases progeny adaptation at the cost of defense. These findings are consistent with an antagonistic relationship between plant immunity and plant growth and development. Warm ambient temperatures induce early flowering in many other plants, including rice. It would be of interest to determine whether the same epigenetic mechanism and SGS3 degradation are utilized by other plants to execute heat responses and thermomemory. Furthermore, how HTT5 coordinates the heat-induced changes in development and immunity is of interest. We observed that these thermomemory phenotypes mostly lasted for 2 generations. This ensures plants to reset responses to ever changing environments.
We propose that H3K27me3 demethylation and HSFA2 activation might be key effectors downstream of heat sensing in plants. How sense heat is still an important topic to be addressed. It is difficult to define the primary heat sensor(s), given that heat simultaneously poses a stress to almost all macromolecules and all organelles in cells. Although several putative heat sensors have been proposed, including a cyclic nucleotide gated calcium channel (CNGC), two unfolded protein sensors, and H2A.Z-containing nucleosomes,3 it is unclear whether these potential sensors-mediated mechanisms are also responsible for the H3K27me3 demethylation-mediated thermomemory. It is possible that plants sense heat through different organelles during different phases of the response. Systematic screening for factors regulating transgenerational thermomemory will shed light on this issue, and might provide new knowledge and technology to improve thermotolerance in crops.
Materials and methods
Plant materials and growth conditions
All genetic stocks used in this paper are in Columbia background. sgs3–12,19 p35S::SGS3-GFP,10 hsfa2,42 pHSFA2::HSFA2-YFP,43 brm-1, ref6–1, ref6–1brm-1, ref6–1 pREF6::REF6-GFP, brm-1 ProBRM::BRM-GFP27 and MIM480(+)8 were described previously. The sgip1 mutant (SALK_017126C) was obtained from the Arabidopsis Biological Resource Center. Homozygous T-DNA insertion mutants were identified by PCR-based genotyping. The progeny of Col 30 °C♀×Col 22 °C♂and Col 22 °C♀×Col 22 °C♂were derived from reciprocal crosses between 22 °C-grown Col and 30 °C-grown Col. After crossing, the parent plants were all kept at 22 °C and the seeds were harvested for further analysis.
The seeds were plated on 1/2 Murashige and Skoog (MS) agar medium and stratified at 4 °C in darkness for 4 day, and then moved to a greenhouse and germinated at 22 °C for 1 week. The seedlings were transplanted to the soil in plastic pots and grown at 22 °C under long-day conditions (16-h light/8-h dark) and 50–70% relative humidity as previously described.10 For heat treatment, 10-day-old seedlings grown at 22 °C were transferred into a growth chamber and grown at 30 °C for 2 weeks with 16-h light/8-h dark cycles and 50–70% relative humidity as previously described.10 All flowering plants were moved back to 22 °C to produce seeds (Supplementary information, Figure S1a). All air-dried seeds were collected and kept at 4 °C.
Flowering time assessment
Wild-type and mutant plants were grown side by side in soil at 22 °C or 30 °C. To compare flowering times among different genotypes, total rosette and cauline leaves were counted when the plants flowered. At least fifteen plants were counted for each line and the analysis was repeated three times independently.
Plasmid constructs and plant transformation
The full-length genomic sequences of SGIP1 and HSFA2 were cloned into pCambia1300 to generate p35S::SGIP1 and p35S::HSFA2–4MYC. The siR480(+)/siR255-resistant version of HTT5 was also cloned into pCambia1300 to generate p35S::rHTT5–4MYC. Two sgRNAs were designed and fused to pCAMBIA1300-pYAO:Cas944 to delete a region (+441 ~ +544 from the first codon) containing CTCTGYTY motifs in the intron of HSFA2. Two sgRNAs targeting the exon of HTT5 were fused to pCAMBIA1300-pYAO:Cas9 to mutate HTT5. All the constructs were introduced into Agrobacterium tumefaciens strain GV3101 to transform Col plants, leading to the generation of more than 15 transgenic plants. For selection of transgenic plants, the seeds were sterilized and germinated on 1/2 MS agar medium containing 25 mg/L hygromycin. All primer sequences used for cloning can be found in Supplementary information, Table S2.
RNA analysis
Total RNAs were extracted using TRIzol reagent from the fourth and fifth true leaves from 24-day-old plants. RNA was quantified using NanoDrop, and approximately 1 µg total RNA was converted into cDNA using HiScript Q RT SuperMix (Vazyme, R123–01) for quantitative real-time PCR (qRT-PCR) according to the manufacturer instructions. 2 μL of the resulting 20 μL cDNAs was used for qRT-PCR analyses with the primers listed in Supplementary Table 2. The qRT-PCR reactions were performed using a CFX96 Real-time PCR Instrument (Bio-Rad) with SYBR® Premix Ex Taq™ (Takara, RR420A). Three biological repeats were analyzed. qRT-PCR data were normalized to the ACTIN2 signals.
To analyze small RNAs, 20–30 μg of total RNAs was separated by electrophoresis on 15% polyacrylamide/7 M urea denaturing gels and transferred to nylon membrane (GE Health) for 1 h at 3.3 mA/cm2 using semi-dry transfer unit (Bio-Rad). After cross-linking by UV irradiation, the membrane was hybridized in PerfectHybTM Plus hybridization buffer (Sigma-Aldrich, H7033) with biotin-labeled DNA probes complementary to the siRNA or miRNA sequences at 42 °C overnight. Detection of immobilized nucleic acids was performed using Nucleic Acid Detection Module Kit (Thermo Fisher, 89880). After stripping the probes, the same blots were reprobed with U6 probe that serves as a loading control.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed as described.45 Briefly, 2 g four-week-old 22 °C- or 30 °C-grown seedlings were harvested and cross-linked with 1% formaldehyde for 10 min under vacuum. For validation of REF6 binding, 0.5 g flowers from 22 °C- or 30 °C-grown plants were used. Cross-linking was stopped by adding glycine. The seedlings were rinsed with water, and then ground into fine powder in liquid nitrogen. Chromatin was isolated and sheared into 200–500 bp fragments by sonication. The sonicated chromatin was incubated with antibodies against GFP (Abcam, ab290) or H3K27me3 (Millipore, 07–449) overnight at 4 °C. Immunocomplexes were precipitated, washed and eluted and the cross-links were reversed at 65 °C overnight. Precipitated DNA was then recovered with the Dr.GenTLETM precipitation carrier (Takara, 9094) according to the manufacturer’s instructions. Background control ChIP was performed using intergenic regions without H3K27me3 mark to determine the ChIP specificity. ChIP-qPCR was performed with three replicates and relative fold enrichments over the background were shown. ChIP experiments were performed independently at least three times with similar results. Sequences of the primers used for ChIP-qPCR are listed in Supplementary information, Table S2.
Electrophoretic mobility shift assays (EMSA)
The full-length coding sequence of HSFA2 was cloned into pET32a and expressed in E. coli Rosetta (DE3). The His-tagged HSFA2 fusion protein was purified. GST and GST-REF6C fusion proteins were expressed and purified as described.28 DNA fragments were end-labeled with Cy5. 200 ng His or His-HSFA2 protein was supplemented with 10 ng Cy5-labeled DNAs and incubated in the binding buffer (10 mM Tris-HCl, pH 7.6, 25 mM KCl, 2.5 mM MgCl2, 1 mM EDTA, 1 mM DTT, and 10% glycerol) at 25 °C for 30 min. For REF6 binding activity detection, 200 ng protein was mixed with 10 ng Cy5-labeled DNAs in the binding buffer (25 mM Tris-HCl, 100 mM NaCl, 2.5 mM MgCl2, 0.1% CA-630, 10% glycerol, 1 µM ZnSO4, and 1 mM DTT, pH 8.0) and incubated on ice for 1 h. For competition assays, 50- or 100-fold non-labeled competitor DNA was added in the reaction 10 min before the addition of the cy5-labeled probe. The reaction mixtures were resolved on 4.5% native polyacrylamide gels in 0.5× TBE at 100 V for 1–2 h on ice in dark. Gels were directly scanned using FLA 2000 (Fuji).
Protein analysis
The anti-SGS3 and anti-SGIP1 polyclonal antibodies were developed by ABclonal® Technology (Wuhan, China). The SGS3ΔN (290–625aa) and full-length CDS of SGIP1 were cloned into pET32a and expressed in Rosetta (DE3). The His-tagged SGS3ΔN and SGIP1 proteins were purified and used as antigens to raise polyclonal antibodies in rabbit, respectively.
To extract total proteins, 0.1 g fifth true leaves from 24-day-old plants were ground into fine power in liquid nitrogen, homogenized in the protein extraction buffer (150 mM Tris-HCl, pH 7.5, 6 M urea, 2% SDS, and 5% β-mercaptoethanol), boiled for 5 min and then centrifuged at 13,200 rpm for 10 min at 4 °C to remove debris. Proteins were resolved on a 4–20% BiofurawTM Precast Gel (Tanon) by electrophoresis and transferred to PVDF membranes at room temperature for 10–20 min at 25 V using Trans-Blot® TurboTM Blotting System (BIO-RAD). Signals were visualized with High-sig ECL Western Blotting Substrate (Tanon, 180–501), and images were captured using the Tanon-5200 Chemiluminescent imaging system (Tanon). Quantification of protein signal was performed using Image J software. Antibodies against the following proteins were used: HSFA2 (1:800 dilution),42 MYC (MILLIPORE, 05–724; 1:2,000 dilution), FLAG (Sigma, F1804; 1:2,000 dilution), SGS3 (1:1,000 dilution), SGIP1 (1:1,000 dilution), GFP (Abcam, ab290; 1:2,000 dilution), ACTIN (CMCTAG, AT0004; 1:2,000 dilution). ACTIN was used as a loading control.
The co-immunoprecipitatin (Co-IP) procedure was described previously.46 Harvested samples were grounded in liquid nitrogen, homogenized in the lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM PMSF and 1× protease inhibitor cocktail), incubated at 4 °C for 30 min, and centrifuged at 10,000 g for 15 min. The supernatant was mixed with prewashed anti-FLAG M2 magnetic beads (Sigma-Aldrich, M8823), incubated at 4 °C for 2 h and washed three times with lysis buffer. The bound proteins were eluted from the affinity beads by boiling for 5 min in 40 μL 2× SDS loading buffer and analyzed by immunoblot.
For cell-free protein degradation assay, total proteins were extracted from 2-week-old seedlings with the extraction buffer (50 mM Tris-MES, pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail) as described previously.47 Samples were incubated at 22 °C or 30 °C in the presence of 100 μM CHX (Sigma-Aldrich, C4859) or MG132 (Sigma-Aldrich, C2211) for various time periods. All protein immunodetection experiments were performed independently three times with similar results.
Yeast two-hybrid assay
The full-length CDS of SGS3 and SGIP1 and domain deletion mutants SGS3Δ220, SGS3ΔZF, SGS3ΔXS, SGS3ΔCC, SGIP1ΔF, and SGIP1ΔFAID were amplified and subcloned into the pDONR207 vector by BP reaction. The inserts were then fused to the GAL4 activation domain in pDEST-GADT7 or GAL4 DNA binding domain in pDEST-GBKT7 by LR reaction. The combinations of bait and prey constructs were co-transformed into yeast strain AH109 and selected on dropout medium without leucine and tryptophan. To assay protein-protein interactions, clones were grown on dropout medium without leucine, histidine and tryptophan containing 2 mM 3-aminotriazole (Sigma-Aldrich, A8056).
Protein ubiquitination assays
The in vitro ubiquitination assay was carried out as described previously.46 SGS3 protein was transiently expressed in N. benthamiana and purified by immunopreciptation using an anti-SGS3 antibody. For the in vitro ubiquitination assay, the reaction mixture (30 μL) containing 50 ng AtE1, 200 ng AtUBC10, 1 μg ubiquitin, 1 μg purified SGIP1 fused with the MBP tag, and 100 ng SGS3 protein was incubated for 1.5 h at 30 °C with agitation. The reaction was stopped by the addition of 4× protein loading buffer and boiling for 5 min. Samples were separated on a 7.5% SDS-PAGE gel and the ubiquitinated SGS3 was detected by immunoblotting with an anti-SGS3 antibody. The mutated SGIP1 protein (SGIP1ΔFbox) was used as a negative control.
In vivo ubiquitination assay by agroinfiltration was performed following the protocol described previously.48 Coinfiltration expression of SGIP1 and SGS3 was performed in N. benthamiana leaves. MG132 was injected into the leaf tissues 12 h before harvest. Total proteins were extracted from leaf tissues and analyzed by immunoblotting with the anti-SGS3, anti-FLAG and anti-GFP antibodies. GFP was co-infiltrated as an internal control and ACTIN was used as a loading control.
Protoplast isolation and transfection
The plasmids PA7-SGS3-GFP and PA7-SGIP1-RFP were constructed and purified with HiSpeed Plasmid Midi Kit (QIAGEN, 12643). The mesophyll protoplasts were isolated from the leaves of 3-week-old Col using the Tape-Arabidopsis Sandwich procedure.49 The protoplasts were transfected with 20 μg purified DNA using PEG solution (40% (w/v) PEG, 0.1 M CaCl2, and 0.2 M mannitol). After incubation at room temperature for 8 mins, W5 buffer (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 2 mM MES, pH 5.7) was added to stop the transfection. The protoplasts were collected and resuspended with W5 buffer, and then transferred into 6-well plates and incubated overnight. YFP and RFP signals were observed with an Olympus FV1000 confocal laser scanning microscope.
Bacterial disease assays and SA measurement
Bacterial disease assays were performed as described.50 Pseudomonas syringae pv. Tomato (Pst) carrying effector avrRpt2 [Pst DC3000(avrRpt2)] was used in this study. Two-week-old plants were shifted from 22 °C to 30 °C or continuously grown at 22 °C for another 1 week before infection. Bacteria were collected by centrifugation and resuspended in sterile water to OD600 = 0.2 (~1 × 108 cfu/ml). Bacterial suspension was further diluted to OD600 = 0.0002 before syringe-infiltration on the fifth true leaves of plants. Water infiltration was performed as mock inoculation. At 0 and 4 days post inoculation (dpi), leaf discs from ten leaves were collected from each genotype, surface-sterilized with 75% ethanol, rinsed in sterile water, and ground in sterile water for bacteria extraction. Bacterial titer was determined by plating a serial dilution of the suspension onto Luria-Marine plates with 100 μg/ml Rifampicin and 50 μg/ml kanamycin and incubated for 2 days at 28 °C, with three biological repeats. Experiments were repeated independently at least three times.
SA was extracted and quantified as previously described.51 0.1 g leaf tissues from plants syringe-infiltrated with water or Pst DC3000(avrRpt2) were ground for SA extraction, with four biological replicates of each genotype. Contents of SA were determined by GC-MS using a labeled internal standard.
Luciferase biomolecular complementation assay
SGS3 and SGIP1 were fused to N- or C-terminus of firefly luciferase and transformed to Agrobacterium strain GV3101. Overnight cultures of Agrobacteria were collected by centrifugation, resuspended in MES buffer (10 mM MgCl2,10 mM MES, 150 μM AS, pH 5.6), mixed with GV3101 expressing P19 to OD600 of 0.5, and incubated at room temperature for 3–4 h before infiltration. Healthy leaves of 3-week-old N. benthamiana were syringe-infiltrated. 2 days later, the LUC activity was assayed with Luciferase Assay Systems (Promega, E1501) and images were captured using the Tanon-5200 Chemiluminescent imaging system (Tanon).
Statistical analysis
For qPCR analyses, ChIP, flowering time assessment, disease assays and SA level detection, significant differences between samples/lines and the corresponding controls were analyzed using a Student’s t-test for pairwise comparisons, one-way or two-way ANOVA analysis with Tukey’s multiple comparison test as specified in the legends. Samples sharing lowercase letters are not significantly different.
References
Pauli, H. et al. Recent plant diversity changes on Europe’s mountain summits. Science 336, 353–355 (2012).
Lobell, D. B., Schlenker, W. & Costa-Roberts, J. Climate trends and global crop production since 1980. Science 333, 616–620 (2011).
Liu, J., Feng, L., Li, J. & He, Z. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 6, 267 (2015).
Tittel-Elmer, M. et al. Stress-induced activation of heterochromatic transcription. PLoS Genet. 6, e1001175 (2010).
Pecinka, A. et al. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22, 3118–3129 (2010).
Ito, H. et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119 (2011).
Lang-Mladek, C. et al. Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol. Plant 3, 594–602 (2010).
Li, S. et al. HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in Arabidopsis. Plant Cell 26, 1764–1780 (2014).
Yu, X. et al. Global analysis of cis-natural antisense transcripts and their heat-responsive nat-siRNAs in Brassica rapa. BMC Plant Biol. 13, 208 (2013).
Zhong, S. H. et al. Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in. Arab. Proc. Natl Acad. Sci. USA 110, 9171–9176 (2013).
Lamke, J. & Baurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18, 124 (2017).
Iwasaki, M. & Paszkowski, J. Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc. Natl Acad. Sci. USA 111, 8547–8552 (2014).
Migicovsky, Z., Yao, Y. & Kovalchuk, I. Transgenerational phenotypic and epigenetic changes in response to heat stress in Arabidopsis thaliana. Plant Signal. Behav. 9, e27971 (2014).
Fukunaga, R. & Doudna, J. A. dsRNA with 5' overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 28, 545–555 (2009).
Kumar, S. V. et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245 (2012).
Huot, B. et al. Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat. Commun. 8, 1808 (2017).
Gangappa, S. N., Berriri, S. & Kumar, S. V. PIF4 coordinates thermosensory growth and immunity in Arabidopsis. Curr. Biol. 27, 243–249 (2017).
Allen, E., Xie, Z., Gustafson, A. M. & Carrington, J. C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 (2005).
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L. & Poethig, R. S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379 (2004).
Karasov, T. L., Chae, E., Herman, J. J. & B, J. Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29, 666–680 (2017).
Deng, Y. et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965 (2017).
Mazzucotelli, E. et al. The E3 ubiquitin ligase gene family in plants: regulation by degradation. Curr. Genom. 7, 509–522 (2006).
Gould, P. D. et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18, 1177–1187 (2006).
Howell, M. D. et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19, 926–942 (2007).
Rajagopalan, R., Vaucheret, H., Trejo, J. & Bartel, D. P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425 (2006).
Nishizawa, A. et al. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547 (2006).
Li, C. et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48, 687–693 (2016).
Cui, X. et al. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 48, 694–699 (2016).
Wang, X. & Moazed, D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science 356, 88–91 (2017).
Laprell, F., Finkl, K. & Muller, J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 356, 85–88 (2017).
Coleman, R. T. & Struhl, G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 356, eaai8236 (2017).
Yu, R., Wang, X. & Moazed, D. Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation. Nature 558, 615–619 (2018).
Klosin, A., Casas, E., Hidalgo-Carcedo, C., Vavouri, T. & Lehner, B. Transgenerational transmission of environmental information in C. elegans. Science 356, 320–323 (2017).
Camacho, J. et al. The memory of environmental chemical exposure in C. elegans is dependent on the jumonji demethylases jmjd-2 and jmjd-3/utx-1. Cell Rep. 23, 2392–2404 (2018).
Yang, H. et al. Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 357, 1142–1145 (2017).
Tao, Z. et al. Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551, 124–128 (2017).
Autran, D. et al. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145, 707–719 (2011).
Chen, M. et al. Maternal temperature history activates Flowering Locus T in fruits to control progeny dormancy according to time of year. Proc. Natl Acad. Sci. USA 111, 18787–18792 (2014).
Chen, M. & Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 360, 1014–1017 (2018).
Zenk, F. et al. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 357, 212–216 (2017).
Whittle, C. A., Otto, S. P., Johnston, M. O. & Krochko, J. E. Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany 87, 650–657 (2009).
Liu, H. C., Liao, H. T. & Charng, Y. Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34, 738–751 (2011).
Lamke, J., Brzezinka, K., Altmann, S. & Baurle, I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 35, 162–175 (2016).
Yan, L. et al. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823 (2015).
Wang, H. et al. Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of polycomb group proteins in association with distinct sets of cis-regulatory elements. PLoS Genet. 12, e1005771 (2016).
You, Q. et al. An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20, 758–769 (2016).
Zhang, H. et al. The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27, 214–227 (2015).
Liu, L., Zhao, Q. & Xie, Q. In vivo ubiquitination assay by agroinfiltration. Methods Mol. Biol. 876, 153–162 (2012).
Wu, F. H. et al. Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009).
Xin, X. F. et al. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539, 524–529 (2016).
Yin, X. et al. Rice copine genes OsBON1 and OsBON3 function as suppressors of broad-spectrum disease resistance. Plant Biotechnol. J. 16, 1476–1487 (2018).
Acknowledgements
We thank Drs. Yee-yung Charng, Hervé Vaucheret, Chenlong Li, Yuhai Cui, Jiawei Wang, Lin Xu, and Yiming Wang for materials and technical assistance; Drs. Jian-Kang Zhu, Xiaoya Chen, and Jonathan Jones for helpful discussion. This project was supported by grants from the National Natural Science Foundation of China (91640114 and 31330061 to Z.H, 31788103 and 91540203 to X.C., 31600207 to J.L. and 31770323 to X.D), Chinese Academy of Sciences (XDB27040201 to Z.H, QYZDY-SSW-SMC022 to X.C.), the National Key Laboratory of Plant Molecular Genetics and the State Key Laboratory of Plant Genomics, the China Postdoctoral Science Foundation (2016M590386 to J.L.), and the Youth Innovation Promotion Association of CAS (2018131 to X.D.).
Author information
Authors and Affiliations
Contributions
J.L., J.Li, X.C., and Z.H. conceived and designed the study. J.L., L.F., X.G. and X.D. performed most of the experiments. Q.L., Y.Z., M.W., and Y.D. performed growth and disease assays. Q.Q., X.D., I.B., Y.H., E.W., X.C., J.L., L.F., and Z.H. developed materials. J.L., X.C., and Z.H. analyzed data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, J., Feng, L., Gu, X. et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res 29, 379–390 (2019). https://doi.org/10.1038/s41422-019-0145-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-019-0145-8
This article is cited by
-
A review of the potential involvement of small RNAs in transgenerational abiotic stress memory in plants
Functional & Integrative Genomics (2024)
-
Revisiting plant stress memory: mechanisms and contribution to stress adaptation
Physiology and Molecular Biology of Plants (2024)
-
Plants and global warming: challenges and strategies for a warming world
Plant Cell Reports (2024)
-
Unveiling the dynamic relationship of viruses and/or symbiotic bacteria with plant resilience in abiotic stress
Stress Biology (2024)
-
The OsSGS3-tasiRNA-OsARF3 module orchestrates abiotic-biotic stress response trade-off in rice
Nature Communications (2023)