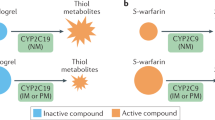
Key Points
-
Substantial progress has been made in the field of pharmacogenomics, with advances in genotyping and sequencing technology, and by the routine collection of DNA samples to study the drug-response phenotype
-
Genetic markers associated with drug toxicity and drug efficacy can be identified by candidate gene, genome-wide association, and next-generation sequencing studies
-
The potential of targeting the right patient with the right drug, and FDA labelling guidance to use pharmacogenetic markers, have provided new impetus to conduct genotype-based randomized clinical trials (RCTs)
-
Prospective approaches using a pharmacogenetic-based strategy with enrichment or adaptive designs are being increasingly used in cardiovascular RCTs
-
Clinical adoption of pharmacogenetics in the practice of cardiovascular medicine will become a reality when a transition has been made from conducting genetic association studies to rigorously performed genotype-based RCTs
Abstract
Consensus practice guidelines and the implementation of clinical therapeutic advances are usually based on the results of large, randomized clinical trials (RCTs). However, RCTs generally inform us on an average treatment effect for a presumably homogeneous population, but therapeutic interventions rarely benefit the entire population targeted. Indeed, multiple RCTs have demonstrated that interindividual variability exists both in drug response and in the development of adverse effects. The field of pharmacogenomics promises to deliver the right drug to the right patient. Substantial progress has been made in this field, with advances in technology, statistical and computational methods, and the use of cell and animal model systems. However, clinical implementation of pharmacogenetic principles has been difficult because RCTs demonstrating benefit are lacking. For patients, the potential benefits of performing such trials include the individualization of therapy to maximize efficacy and minimize adverse effects. These trials would also enable investigators to reduce sample size and hence contain costs for trial sponsors. Multiple ethical, legal, and practical issues need to be considered for the conduct of genotype-based RCTs. Whether pre-emptive genotyping embedded in electronic health records will preclude the need for performing genotype-based RCTs remains to be seen.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Pereira, N. L. & Weinshilboum, R. M. Cardiovascular pharmacogenomics and individualized drug therapy. Nat. Rev. Cardiol. 6, 632–638 (2009).
Wang, L., McLeod, H. L. & Weinshilboum, R. M. Genomics and drug response. N. Engl. J. Med. 364, 1144–1153 (2011).
Pereira, N. L. & Weinshilboum, R. M. The impact of pharmacogenomics on the management of cardiac disease. Clin. Pharmacol. Ther. 90, 493–495 (2011).
Koboldt, D. C., Steinberg, K. M., Larson, D. E., Wilson, R. K. & Mardis, E. R. The next-generation sequencing revolution and its impact on genomics. Cell 155, 27–38 (2013).
Bowton, E. et al. Biobanks and electronic medical records: enabling cost-effective research. Sci. Transl. Med. 6, 234cm3 (2014).
Thorn, C., Klein, T. & Altman, R. PharmGKB: The Pharmacogenomics Knowledge Base. Pharmacogenomics 1015, 311–320 (2013).
MacRae, C. A. Cardiac arrhythmia: in vivo screening in the zebrafish to overcome complexity in drug discovery. Expert Opin. Drug Discov. 5, 619–632 (2010).
Jiang, J. et al. Genome-wide association study for biomarker identification of Rapamycin and Everolimus using a lymphoblastoid cell line system. Front. Genet. 4, 166 (2013).
Volzke, H. et al. Personalized cardiovascular medicine: concepts and methodological considerations. Nat. Rev. Cardiol. 10, 308–316 (2013).
Wang, B., Canestaro, W. J. & Choudhry, N. K. Clinical evidence supporting pharmacogenomic biomarker testing provided in US Food and Drug Administration drug labels. JAMA Intern. Med. 174, 1938–1944 (2014).
Ahmad, T. et al. Charting a roadmap for heart failure biomarker studies. JACC Heart Fail. 2, 477–488 (2014).
Pirmohamed, M. et al. A randomized trial of genotype-guided dosing of warfarin. N. Engl. J. Med. 369, 2294–2303 (2013).
Kimmel, S. E. et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 369, 2283–2293 (2013).
Bergmeijer, T. O. et al. CYP2C19 genotype–guided antiplatelet therapy in ST-segment elevation myocardial infarction patients—rationale and design of the Patient Outcome after primary PCI (POPular) Genetics study. Am. Heart J. 168, 16–22.e1 (2014).
Takeuchi, F. et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 5, e1000433 (2009).
Liggett, S. B. et al. A polymorphism within a conserved β1-adrenergic receptor motif alters cardiac function and β-blocker response in human heart failure. Proc. Natl Acad. Sci. 103, 11288–11293 (2006).
SEARCH Collaborative Group. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N. Engl. J. Med. 359, 789–799 (2008).
Daneshjou, R. et al. Genetic variant in folate homeostasis is associated with lower warfarin dose in African Americans. Blood 124, 2298–2305 (2014).
Myocardial Infarction Genetics Consortium Investigators. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N. Engl. J. Med. 371, 2072–2082 (2014).
Bollag, G. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599 (2010).
Aithal, G. P., Day, C. P., Kesteven, P. J. L. & Daly, A. K. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 353, 717–719 (1999).
Rost, S. et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 427, 537–541 (2004).
Zhang, J. E. et al. Effects of CYP4F2 genetic polymorphisms and haplotypes on clinical outcomes in patients initiated on warfarin therapy. Pharmacogenet. Genomics 19, 781–789 (2009).
Ioannidis, J. P. To replicate or not to replicate: the case of pharmacogenetic studies: have pharmacogenomics failed, or do they just need larger-scale evidence and more replication? Circ. Cardiovasc. Genet. 6, 413–418 (2013).
Aslibekyan, S., Claas, S. A. & Arnett, D. K. To replicate or not to replicate: the case of pharmacogenetic studies establishing validity of pharmacogenomic findings: from replication to triangulation. Circ. Cardiovasc. Genet. 6, 409–412 (2013).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Weeke, P. et al. Exome sequencing implicates an increased burden of rare potassium channel variants in the risk of drug-induced long QT interval syndrome. J. Am. Coll. Cardiol. 63, 1430–1437 (2014).
Ashley, E. A. et al. Clinical assessment incorporating a personal genome. Lancet 375, 1525–1535 (2010).
Biesecker, L. G. & Green, R. C. Diagnostic clinical genome and exome sequencing. N. Engl. J. Med. 370, 2418–2425 (2014).
Green, R. C. et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 15, 565–574 (2013).
Goldstein, D. B. et al. Sequencing studies in human genetics: design and interpretation. Nat. Rev. Genet. 14, 460–470 (2013).
Scriver, C. R. & Childs, B. (eds) Garrod's Inborn Factors in Disease (Oxford University Press, 1989).
Johnson, J. A. et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin. Pharmacol. Ther. 90, 625–629 (2011).
Paré, G. et al. Genetic determinants of dabigatran plasma levels and their relation to bleeding. Circulation 127, 1404–1412 (2013).
Mega, J. L. et al. Cytochrome P-450 polymorphisms and response to clopidogrel. N. Engl. J. Med. 360, 354–362 (2009).
Mega, J. L. et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 304, 1821–1830 (2010).
Wang, L. et al. Human thiopurine S-methyltransferase pharmacogenetics: variant allozyme misfolding and aggresome formation. Proc. Natl Acad. Sci. USA 102, 9394–9399 (2005).
Lennard, L., Van Loon, J. A. & Weinshilboum, R. M. Pharmacogenetics of acute azathioprine toxicity: relationship to thiopurine methyltransferase genetic polymorphism. Clin. Pharmacol. Ther. 46, 149–154 (1989).
Van Loon, J. A. & Weinshilboum, R. M. Human lymphocyte thiopurine methyltransferase pharmacogenetics: effect of phenotype on 6-mercaptopurine-induced inhibition of mitogen stimulation. J. Pharmacol. Exp. Ther. 242, 21–26 (1987).
Liang, J. J. et al. TPMT genetic variants are associated with increased rejection with azathioprine use in heart transplantation. Pharmacogenet. Genomics 23, 658–665 (2013).
Price, M. J. et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. JAMA 305, 1097–1105 (2011).
Collet, J.-P. et al. Bedside monitoring to adjust antiplatelet therapy for coronary stenting. N. Engl. J. Med. 367, 2100–2109 (2012).
Martin, A. M. et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc. Natl Acad. Sci. 101, 4180–4185 (2004).
Mallal, S. et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 359, 727–732 (2002).
Hetherington, S. et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet 359, 1121–1122 (2002).
Hughes, A. R. et al. Association of genetic variations in HLA-B region with hypersensitivity to abacavir in some, but not all, populations. Pharmacogenomics 5, 203–211 (2004).
Phillips, E. J. et al. Clinical and immunogenetic correlates of abacavir hypersensitivity. AIDS 19, 979–981 (2005).
Mallal, S. et al. HLA-B*5701 screening for hypersensitivity to abacavir. N. Engl. J. Med. 358, 568–579 (2008).
Ramsey, B. W. et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N. Engl. J. Med. 365, 1663–1672 (2011).
US Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labeling [online], (2015).
Scott, S. A. et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 94, 317–323 (2013).
Teutsch, S. M. et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genet. Med. 11, 3–14 (2009).
Roberts, J. D. et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet 379, 1705–1711 (2012).
Lala, A. et al. Genetic testing in patients with acute coronary syndrome undergoing percutaneous coronary intervention: a cost-effectiveness analysis. J. Thromb. Haemost. 11, 81–91 (2013).
Kazi, D. S. et al. Cost-effectiveness of genotype-guided and dual antiplatelet therapies in acute coronary syndrome. Ann. Intern. Med. 160, 221–232 (2014).
Urban, T. J. & Goldstein, D. B. Pharmacogenetics at 50: genomic personalization comes of age. Sci. Transl. Med. 6, 220ps1 (2014).
Simon, R. M., Paik, S. & Hayes, D. F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl Cancer Inst. 101, 1446–1452 (2009).
Douillard, J.-Y. et al. Panitumumab–FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 369, 1023–1034 (2013).
Wallentin, L. et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet 376, 1320–1328 (2010).
Sorich, M. J., Vitry, A., Ward, M. B., Horowitz, J. D. & McKinnon, R. A. Prasugrel vs. clopidogrel for cytochrome P450 2C19-genotyped subgroups: integration of the TRITON-TIMI 38 trial data. J. Thromb. Haemost. 8, 1678–1684 (2010).
US National Library of Medicine. ClinicalTrials.gov [online], (2015).
US National Library of Medicine. ClinicalTrials.gov [online], (2014).
Maitournam, A. & Simon, R. On the efficiency of targeted clinical trials. Stat. Med. 24, 329–339 (2005).
Freidlin, B., Korn, E. L. & Gray, R. Marker Sequential Test. (MaST) design. Clin. Trials 11, 19–27 (2014).
Mandrekar, S. J. & Sargent, D. J. Clinical trial designs for predictive biomarker validation: theoretical considerations and practical challenges. J. Clin. Oncol. 27, 4027–4034 (2009).
Le Tourneau, C. et al. Randomised proof-of-concept phase II trial comparing targeted therapy based on tumour molecular profiling vs conventional therapy in patients with refractory cancer: results of the feasibility part of the SHIVA trial. Br. J. Cancer 111, 17–24 (2014).
Kim, E. S. et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 1, 44–53 (2011).
Barker, A. D. et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin. Pharmacol. Ther. 86, 97–100 (2009).
Korn, E. L. & Freidlin, B. Outcome-adaptive randomization: is it useful? J. Clin. Oncol. 29, 771–776 (2011).
Abrams, J. et al. National Cancer Institute's Precision Medicine Initiatives for the new National Clinical Trials Network. Am. Soc. Clin. Oncol. Educ. Book 34, 71–76 (2014).
CIBIS-II Investigators and Committees. The Cardiac Insufficiency Bisoprolol Study II (CIBIS-II): a randomised trial. Lancet 353, 9–13 (1999).
Packer, M. et al. The effect of carvedilol on morbidity and mortality in patients with chronic heart failure. N. Engl. J. Med. 334, 1349–1355 (1996).
Hjalmarson, Å. et al. Effects of controlled-release metoprolol on total mortality, hospitalizations, and well-being in patients with heart failure: Tte metoprolol CR/XL randomized intervention trial in congestive heart failure (MERIT-HF). JAMA 283, 1295–1302 (2000).
Beta-Blocker Evaluation of Survival Trial Investigators. A trial of the beta-blocker bucindolol in patients with advanced chronic heart failure. N. Engl. J. Med. 344, 1659–1667 (2001).
Moore, J. D., Mason, D. A., Green, S. A., Hsu, J. & Liggett, S. B. Racial differences in the frequencies of cardiac β1-adrenergic receptor polymorphisms: analysis of c145A>G and c1165G>C. Hum. Mutat. 14, 271 (1999).
Tirona, R. G., Leake, B. F., Merino, G. & Kim, R. B. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J. Biol. Chem. 276, 35669–35675 (2001).
Pasanen, M. K., Neuvonen, M., Neuvonen, P. J. & Niemi, M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet. Genomics 16, 873–879 (2006).
Ramsey, L. B. et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin. Pharmacol. Ther. 96, 423–428 (2014).
Danik, J. S. et al. Lack of association between SLCO1B1 polymorphisms and clinical myalgia following rosuvastatin therapy. Am. Heart J. 165, 1008–1014 (2013).
Brunham, L. R. et al. Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. Pharmacogenomics J. 12, 233–237 (2012).
Voora, D. et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J. Am. Coll. Cardiol. 54, 1609–1616 (2009).
US National Library of Medicine. ClinicalTrials.gov [online], (2014).
Budnitz, D. S., Shehab, N., Kegler, S. R. & Richards, C. L. Medication use leading to emergency department visits for adverse drug events in older adults. Ann. Intern. Med. 147, 755–765 (2007).
US Food and Drug Administration. Coumadin (warfarin sodium) tablets label [online], (2011).
Go, A. S. et al. Heart disease and stroke statistics—2014 update: a report from the American Heart Association. Circulation 129, e28–e292 (2014).
de Morais, S. M. et al. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J. Biol. Chem. 269, 15419–15422 (1994).
De Morais, S. M. et al. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol. Pharmacol. 46, 594–598 (1994).
Scott, S. A. et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450–452C19 (CYP2C19) genotype and clopidogrel therapy. Clin. Pharmacol. Ther. 90, 328–332 (2011).
Brandt, J. T. et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J. Thromb. Haemost. 5, 2429–2436 (2007).
Holmes, M. V., Perel, P., Shah, T., Hingorani, A. D. & Casas, J. P. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA 306, 2704–2714 (2011).
Umemura, K., Furuta, T. & Kondo, K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J. Thromb. Haemost. 6, 1439–1441 (2008).
US Food and Drug Administration. FDA drug safety communication: reduced effectiveness of Plavix (clopidogrel) in patients who are poor metabolizers of the drug [online], (2014).
Holmes, D. R. Jr et al. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J. Am. Coll. Cardiol. 56, 321–341 (2010).
Levine, G. N. et al. 2011 ACCF/AHA/SCAI guideline for percutaneous coronary intervention: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Society for Cardiovascular Angiography and Interventions. J. Am. Coll. Cardiol. 58, e44–e122 (2011).
US National Library of Medicine. ClinicalTrials.gov [online], (2013).
Mrazek, D. A. & Lerman, C. Facilitating clinical implementation of pharmacogenomics. JAMA 306, 304–305 (2011).
Rothstein, M. A. & Epps, P. G. Ethical and legal implications of pharmacogenomics. Nat. Rev. Genet. 2, 228–231 (2001).
Kohane, I. S. Using electronic health records to drive discovery in disease genomics. Nat. Rev. Genet. 12, 417–428 (2011).
Xu, H. et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J. Am. Med. Inform. Assoc. 18, 387–391 (2011).
Rasmussen-Torvik, L. J. et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin. Pharmacol. Ther. 96, 482–489 (2014).
Daly, A. K. Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 11, 241–246 (2010).
Altman, R. B., Whirl-Carrillo, M. & Klein, T. E. Challenges in the pharmacogenomic annotation of whole genomes. Clin. Pharmacol. Ther. 94, 211–213 (2013).
Acknowledgements
Supported in part by Mayo Transplant Scholarly Award (N.L.P.), U01 GM61388 (The Pharmacogenetics Research Network). The TAILOR-PCI study is funded in part by the Mayo Clinic Centre for Individualized Medicine, and the Mayo Clinic Division of Cardiology. We thank Ms Luanne Wussow (Mayo Clinic, Rochester, MN, USA) for her assistance with the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
N.L.P. researched data for the article. N.L.P., D.J.S., and C.S.R. provided substantial contributions to discussion of the content. N.L.P., D.J.S., M.E.F., and C.S.R. wrote and reviewed/edited the article before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Pereira, N., Sargent, D., Farkouh, M. et al. Genotype-based clinical trials in cardiovascular disease. Nat Rev Cardiol 12, 475–487 (2015). https://doi.org/10.1038/nrcardio.2015.64
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrcardio.2015.64
This article is cited by
-
Next-Generation Sequencing of CYP2C19 in Stent Thrombosis: Implications for Clopidogrel Pharmacogenomics
Cardiovascular Drugs and Therapy (2021)
-
Cholesterol metabolism in mice models of genetic hypercholesterolemia
Journal of Physiology and Biochemistry (2020)
-
Antiplatelet Therapy for Secondary Prevention of Vascular Disease Complications
Current Atherosclerosis Reports (2017)
-
Personalizing Antiplatelet Therapies for Acute Coronary Syndrome (ACS) in Patients Undergoing Percutaneous Coronary Intervention (PCI): Are They Cost-effective?
Cardiovascular Drugs and Therapy (2017)
-
Pharmacogenetics in Cardiovascular Medicine
Current Genetic Medicine Reports (2016)