Abstract
The deregulation of miR-101 and DNMT3a has been implicated in the pathogenesis of multiple tumor types, but whether and how miR-101 silencing and DNMT3a overexpression contribute to lung tumorigenesis remain elusive. Here we show that miR-101 downregulation associates with DNMT3a overexpression in lung cancer cell lines and patient tissues. Ectopic miR-101 expression remarkably abrogated the DNMT3a 3′-UTR luciferase activity corresponding to the miR-101 binding site and caused an attenuated expression of endogenous DNMT3a, which led to a reduction of global DNA methylation and the re-expression of tumor suppressor CDH1 via its promoter DNA hypomethylation. Functionally, restoration of miR-101 expression suppressed lung cancer cell clonability and migration, which recapitulated the DNMT3a knockdown effects. Interestingly, miR-101 synergized with decitabine to downregulate DNMT3a and to reduce DNA methylation. Importantly, ectopic miR-101 expression was sufficient to trigger in vivo lung tumor regression and the blockage of metastasis. Consistent with these phenotypes, examination of xenograft tumors disclosed an increase of miR-101, a decrease of DNMT3a and the subsequent DNA demethylation. These findings support that the loss or suppression of miR-101 function accelerates lung tumorigenesis through DNMT3a-dependent DNA methylation, and suggest that miR-101-DNMT3a axis may have therapeutic value in treating refractory lung cancer.
Similar content being viewed by others
Main
Owing to a high propensity for recurrence and a high rate of metastasis at the advanced stages,1, 2, 3 lung cancer remains the leading cause of cancer-related mortality. DNA methylation is a major epigenetic rule controlling chromosomal stability and gene expression.4, 5 It is under control of DNA methyltransferases (DNMTs), whose overexpression in lung cancer cells predicts worse outcomes.6, 7 It is postulated that DNMT overexpression induces DNA hypermethylation and silencing of tumor suppressor genes (TSGs), leading to an aggressive lung cancer. Indeed, enforced expression of DNMT1 or DNMT3a increases DNA methylation, while the abolition of DNMT expression by genetic depletion, microRNAs (miRs) or small molecules reduces genome-wide and gene-specific DNA methylation and restores TSG expression.8, 9, 10, 11, 12, 13 As TSGs are the master controllers for cell multiplicity and their silencing predicts poor prognosis,14, 15 TSG re-expression via promoter DNA hypomethylation inhibits cell proliferation and induces cell differentiation.13, 16 Thus, DNMT gene abundance could serve as a target for anticancer therapy, but how DNMT upregulation occurs in lung cancer is incompletely understood.
MiRs are small non-coding RNAs that crucially regulate target gene expression. Up to 30% of all protein-coding genes are predicted to be targeted by miRs,17, 18 supporting the key roles of miRs in controlling cell fate.19, 20, 21, 22 Research is showing that certain miRs are frequently dysregulated in cancers, including lung cancer.7, 23, 24 As miR targets can promote or inhibit cancer cell expansion, miRs have huge potential for acting as bona fide oncogenes (i.e., miR-21) or TSGs (i.e., miR-29b).7, 25 We and others demonstrated that the levels of DNMT1 or DNMT3a or DNMT3b are regulated by miR-29b, miR-148, miR-152 or miR-30c,7, 13, 26, 27 and overexpression of these miRs results in DNA hypomethylation and TSG reactivation with the concurrent blockage of cancer cell proliferation.7, 13 These findings underscore the importance of miRs as epigenetic modulators and highlight their therapeutic applications.
MiR-101 is frequently silenced in human cancers28, 29, 30, 31 and, importantly, exhibits antitumorigenic properties when overexpressed. Mechanistically, miR-101 inactivation by genomic loss causes the overexpression of EZH2, a histone methyltransferase, via 3′-UTR targeting, which is followed by histone hypermethylation and aggressive tumorigenesis.29, 30, 32 However, whether and how miR-101 silencing contributes to DNA hypermethylation patterning in lung cancer is unclear. In this study, we explore the role of miR-101 in regulating DNMT3a expression and the impacts of miR-101-DNMT3a nexus on lung cancer pathogenesis. We showed that the expression of miR-101 and DNMT3a was negatively correlated in lung cancer. We presented evidence that ectopic miR-101 expression decreased DNMT3a levels, reduced global DNA methylation and upregulated CDH1 via its promoter DNA demethylation. The biological significance of miR-101-mediated DNA hypomethylation and CDH1 re-expression was evident by its inhibition of lung tumor cell growth in vitro and in vivo. Thus, our findings mechanistically and functionally link miR-101 silencing to DNA hypermethylation in lung cancer cells.
Results
MiR-101 levels are inversely correlated with DNMT3a expression in lung cancer
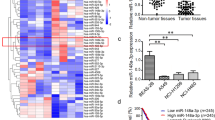
Although miR-101 deregulation and DNMT3a overexpression are implicated in the pathogenesis of multiple tumor types, whether and how these aberrations cooperatively promote lung tumorigenesis are unclear. Using quantitative PCR (qPCR), we initially evaluated the expression of miR-101 and DNMT3a in fresh-frozen matched non-small cell lung cancer (NSCLC; non-smoker) and adjacent non-tumor lung tissues (n=30), and found that DNMT3a expression was 2.9±0.8 folds higher (P=0.032) in tumors than in normal tissues (Figure 1a, left). In contrast, miR-101 was primarily expressed in normal tissues with a 46% reduction in tumors (P=0.044) (Figure 1a, right). We then assessed DNMT3a and miR-101 expression by qPCR in eight NSCLC and two SCLC cell lines. Consistent with the results from fresh-frozen tissues, DNMT3a levels were highly elevated, whereas miR-101 expression was significantly downregulated, in lung cancer cells, when compared with normal lung fibroblasts MRC-5 (Figure 1b). We further examined miR-101 and DNMT3a expression in human NSCLC tissue microarrays (TMAs) using immunohistochemistry (IHC). The stained TMAs were quantified using the modified Histoscores (H-scores), and the representative images of no staining (H-score, 0), weak staining (H-score, 0–100), moderate staining (H-score, 100–200) and strong staining (H-score, 200–300) of DNMT3a and miR-101 were shown in Figure 1c. These experiments revealed that miR-101 expression primarily occurred in normal tissues and was significantly decreased in patient tissues with lower levels at later stages. In contrast, all the tumors displayed moderate or higher DNMT3a expression as compared with normal tissues (Figure 1d). These findings demonstrate that DNMT3a is overexpressed, whereas miR-101 is downregulated, in lung cancer cells.
DNMT3a and miR-101 are negatively correlated in human lung cancer patient tissues and cell lines. (a) qPCR showing the RNA expressions of miR-101 and DNMT3a in fresh-frozen human lung cancer tissues (normal, n=30; tumor, n=30). (b) qPCR showing the expressions of miR-101 and DNMT3a in human lung cancer cell lines (n=10). qPCR results in cancer cell lines were normalized to that in human normal lung fibroblast MRC-5. (c) TMAs of human NSCLC stained with the DNMT3a (IHC) and miR-101 (ISH). Representative micrographs for each group were shown (magnification × 200). (d) Quantification of miR-101 or DNMT3a of ISH or IHC-stained TMA slides from c by H-score. The graphs indicate the gene expression levels at the different stages of lung cancer. Error bars represent±S.E.; using one-way ANOVA followed by Dunnett’s test; ***P<0.0001 versus the normal lung tissue group. (e) Correlation of miR-101 and DNMT3a in fresh-frozen human lung cancer tissues (n=30). Total RNA was isolated using miRNeasy Mini Kit (Qiagen). (f) Correlation of miR-101 and DNMT3a expression in human lung cancer cell lines (n=10). (g) Correlation of miR-101 and DNMT3a in lung cancer TMAs. For e, f and g, R: Pearson correlation coefficients; R2: the goodness of fit. Statistical significance was calculated by Pearson correlation coefficients
To address whether DNMT3a and miR-101 have potential in-parallel relationship, we analyzed DNMT3a and miR-101 expression using Pearson correlation coefficient and found that DNMT3a upregulation was accompanied by miR-101 downregulation, whereas lower DNMT3a was seen in patients carrying higher miR-101 in fresh-frozen patient tissues (Figures 1e, P=0.029, R2=0.16), cell lines (Figures 1f, P=0.042, R2=0.5) and IHC-stained TMAs (Figures 1g, P<0.001, R2=0.53), indicating a negative correlation between DNMT3a and miR-101 expression. Notably, we also measured DNMT1 and DNMT3b expression, and examined their correlation with miR-101 in lung cancer patient tissues and cell lines. However, miR-101 levels did not show a significant correlation with DNMT1 or DNMT3b expression (Supplementary Figure 1). Thus, DNMT3a overexpression and miR-101 silencing in lung cancer cells may cooperatively modulate lung tumorigenesis.
MiR-101 represses DNMT3a by direct targeting its 3′-UTR
Given the negative roles of miRs in regulating their targets and the inverse correlation of miR-101 and DNMT3a in lung cancer, we posited that miR-101 deregulation may be, at least partially, responsible for abnormal DNMT3a expression. To this end, miR-101 or scramble was introduced into human NSCLC H1975 or H1299, or mouse Lewis lung carcinoma (LLC) cells. qPCR revealed miR-101 upregulation in cells transfected with miR-101, including 5.8±0.6 folds (P<0.01) in H1975, 7.4±0.9 folds (P<0.01) in H1299 and 4.1±0.5 folds (P<0.01) in LLC (Figure 2a), with a significant decrease of DNMT3a mRNA expression by 77±8% (P<0.01) in H1975, 85±9% (P<0.01) in H1299 and 64±7% (P<0.05) in LLC cells (Figure 2b). Western blotting identified a significant reduction of DNMT3a protein levels in miR-101-expressing cells (Figure 2c). Further, EZH2, a known miR-101 target,29 was used as a positive control. Consistent with the previous report,33 enforced miR-101 expression greatly diminished EZH2 protein expression (Figure 2c). Thus, DNMT3a overexpression in lung cancer cells may result from the loss or suppression of miR-101.
DNMT3a is a direct target of miR-101. (a) qPCR showing the levels of miR-101 expression in H1975 or H1299, or LLC cells transfected with miR-101 or scramble for 48 h. The data are represented as mean±S.D. from three independent experiments, **P<0.01. (b) qPCR to measure the endogenous DNMT3a expression in H1975 or H1299, or LLC cells transfected with miR-101 or scramble for 48 h. The data are represented as mean±S.D. from three independent experiments, *P<0.05, **P<0.01. (c) Western blot analysis for the change of DNMT3a and EZH2 protein expression in H1975 or H1299, or LLC cells transfected with miR-101 or scramble for 48 h. (d) Diagram showing the complementary site of miR-101 on DNMT3a 3′-UTR. (e) Co-transfection of H1975 or H1299 cells with scramble or miR-101 plus firefly luciferase fused with DNMT3a 3′-UTR. Luciferase activity was measured at 48 h after transfection, and the relative ratio of the activity in the miR-101 group to that in the control scramble group is presented as the mean±S.D. from three independent experiments, *P<0.05, **P<0.01
To determine whether miR-101 suppresses DNMT3a through a process triggered by the interactions between miR-101 and DNMT3a 3′-UTR, by using miR target prediction algorithm TargetScan, we identified a sequence that matches to the miR-101 seed on DNMT3a 3′-UTR (Figure 2d). We then obtained a reporter plasmid containing firefly luciferase fused with DNMT3a 3′-UTR sequence of miR-101 binding site and cotransfected the reporter plasmid with miR-101 or scramble in H1975 or H1299 cells for 48 h. As shown in Figure 2e, ectopic miR-101 expression caused a remarkable decrease of luciferase activity driven by DNMT3a 3′-UTR in H1975 (72±7%, P<0.05) or H1299 (81±9%, P<0.01) cells. When H1975 or H1299 cells were transfected with miR-101 or scramble, the cells were collected at different time points. As shown in Supplementary Figures 2a and b, in a time-dependent manner, miR-101 was upregulated, but DNMT3a was decreased in miR-101-transfected cells. In contrast, no obvious changes of DNMT1 and DNMT3a expression were seen in the presence of miR-101 (Supplementary Figures 2c and d), supporting the observation that no putative miR-101 binding sites were identified in DNMT1 or DNMT3b 3′-UTR (not shown). These data suggest that DNMT3a is a direct target of miR-101 in lung cancer cells.
Enforced miR-101 expression reduces global DNA methylation and restores CDH1 expression via promoter DNA hypomethylation
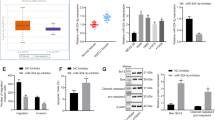
To address whether miR-101 deregulation affects DNA methylation status in lung cancer cells, the genomic DNA was extracted from miR-101-expressing cells and subjected to Dotblot using 5-methylcytosine (5mC) antibody.34 We first used the serial dilution of standard unmethylated (C) or methylated (5mC) and demonstrated that Dotblot is sufficient to detect the small change of 5mC content in genomic DNA (not shown). The equal DNA loading was also confirmed by methylene blue staining. As shown in Figures 3a and b, the representative images of Dotblot indicated a significant reduction of global DNA methylation, which was verified by the quantification of dot intensities in miR-101-transfected H1975 (46±6%, P<0.05), H1299 (64±7%, P<0.05) or LLC (56±7%, P<0.05) cells.
MiR-101 expression induces DNA hypomethylation. (a) The genomic DNA was prepared from H1975 or H1299, or LLC cells transfected with miR-101 or scramble for 48 h and subjected to Dotblot using 5mC antibody. Representative images are shown. Bottom: equal DNA loading was verified by staining the membranes with 0.2% methylene blue. (b) Graphs are the quantification of Dotblot intensity reported in a. The data are represented as mean±S.D. from three independent experiments, *P<0.05. (c) qPCR analysis for the expression of tumor suppressor CDH1 in H1975 or H1299, or LLC cells transfected with miR-101 or scramble for 48 h. The data are represented as mean±S.D. from three independent experiments, *P<0.05, **P<0.01. (d) Bisulfite analysis for the change of DNA methylation in CDH1 gene promoter in H1299 cells transfected with miR-101 or scramble for 48 h. Upper: distribution of CpG dinucleotides is indicated as small vertical lines. Numbers refer to position relative to the transcriptional starting site (TSS) of CDH1; lower: open circles indicate unmethylated CpG sites, filled circles indicate methylated CpG sites. Results of 10 clones are presented
Cadherin 1 (CDH1) is a cell–cell adhesion molecule and functions as a metastasis suppressor in lung cancer.35 It is epigenetically silenced36 and its downregulation associates with poor prognosis in NSCLC.37, 38 As miR-101 regulates DNMT3a-dependent DNA methylation, we proposed that miR-101 downregulation might mediate CDH1 silencing via promoter DNA hypermethylation. In fact, qPCR disclosed a remarkable increase of CDH1 in miR-101-expressing H1975 (3.6±0.4 folds, P<0.05), H1299 (3.4±0.6 folds, P<0.05) or LLC (4.7±0.6 folds, P<0.01) cells, when compared with scramble (Figure 3c). To dissect the underlying mechanisms, we analyzed CDH1 promoter methylation status using bisulfite sequencing in transfected H1975 cells, and found that a greater than 15% change (97% in scramble versus 81% in miR-101) from hyper- to un-methylated was observed in CDH1 promoter (Figure 3d). Thus, miR-101 silencing contributes to the global and gene-specific DNA methylation in lung cancer cells.
DNMT3a expression positively modulates DNA methylation profile
To further demonstrate that miR-101 regulates DNA methylation through DNMT3a, H1975 or H1299 cells were transfected with DNMT3a expression vector or small interfering RNA (siRNA) or their corresponding control, and the effects of DNMT3a manipulation was assessed at 48 h post transfection. As expected, DNMT3a was significantly upregulated at both RNA and protein levels in the presence of DNMT3a expression vector (Figure 4a). By contrast, DNMT3a expression was greatly decreased after siRNA transfection (Figure 4b). Dotblot unveiled a remarkable increase of global DNA methylation in DNMT3a-overexpressing cells (Figure 4c), while a significant reduction of global DNA methylation after DNMT3a depletion (Figure 4d). Functional assays disclosed that DNMT3a overexpression increased lung cancer cell migration in H1975 (912±65 versus 297±54 μm, P<0.05) or H1299 (955±32 versus 366±76 μm, P<0.05) (Figure 4e), whereas DNMT3a abrogation suppressed lung cancer cell mobility (927±56 versus 315±31 μm, P<0.05 in H1975 and 936±33 versus 365±57 μm, P<0.05 in H1299) (Figure 4f), providing a possible explanation for why DNMT3a overexpression independently predicts poor prognosis in lung cancer. Thus, we conclude that miR-101 indirectly modulates DNA methylation program through DNMT3a regulation in lung cancer cells.
The change of DNMT3a expression positively matches the alterations of DNA methylation levels in lung cancer cells. (a) Western blotting (left) and qPCR analysis (right) for DNMT3a expression in H1975 or H1299 cells transfected with DNMT3a expression or empty vector for 48 h. The data are represented as mean±S.D. from three independent experiments, **P<0.01. (b) Western blotting (left) and qPCR analysis (right) for DNMT3a expression in H1975 or H1299 cells transfected with DNMT3a siRNA or scramble for 48 h. The data are represented as mean±S.D. from three independent experiments, *P<0.05. (c) The genomic DNA was prepared from H1975 or H1299 cells transfected with DNMT3a expression or empty vector for 48 h and subjected to Dotblot using 5mC antibody. Left: representative images are shown; right: graph is the quantification of Dotblot intensity from left. The data are represented as mean±S.D. from three independent experiments, *P<0.05, **P<0.01. (d) The genomic DNA was prepared from H1975 or H1299 cells transfected with DNMT3a siRNA or scramble for 48 h and subjected to Dotblot using 5mC antibody. Left: representative images are shown; right: graph is the quantification of Dotblot intensity from left. The data are represented as mean±S.D. from three independent experiments, *P<0.05, **P<0.01. Note, si-DNMT3a represents DNMT3a siRNA. (e) Wound-healing assays evaluating the change of cell mobility in H1975 or H1299 cells transfected with DNMT3a expression or empty vector. Graph is the quantification of ‘gap closure’. The data are represented as mean±S.D. from three independent experiments, *P<0.05. (f) Wound-healing assays for the measurement of cell migration in H1975 or H1299 cells transfected with DNMT3a siRNA or scramble. Graph is the quantification of ‘gap closure’. The data are represented as mean±S.D. from three independent experiments, *P<0.05
MiR-101 upregulation impairs expansion and migration of lung cancer cells in vitro
Given that CDH1 silencing triggers cell proliferation, we speculated that miR-101-rescued CDH1 expression could inhibit lung cancer cell growth. To demonstrate this, H1975 or H1299 or LLC cells were transfected with miR-101 or scramble; at 6 h post transfection when the cells did not undergo apoptosis, ∼1000 cells were subjected to colony-forming assays. As shown in Figures 5a and b, miR-101 overexpression greatly decreased colony number as compared with scramble, in H1975 (54±7 versus 153±17, P<0.01), H1299 (96±14 versus 269±36, P<0.01) or LLC (42±6 versus 126±14, P<0.01).
Lung cancer cell proliferation and migration are reduced by miR-101 in vitro. (a and b) Colony-forming assays in H1975 or H1299, or LLC cells transfected with miR-101 or scramble for 48 h. Representative images are shown in a. Graphs are the quantification of colony numbers (b). The data are represented as mean±S.D. from three independent experiments, **P<0.01. (c and d) Wound-healing assays for cell mobility in H1975 or H1299 or LLC cells transfected with miR-101 or scramble for 48 h. Representative images are shown in c. Graphs are the quantification of ‘gap closure’ (d). The data are represented as mean±S.D. from three independent experiments, *P<0.05, **P<0.01. (e) Western blotting for detecting the activated caspase-3 in H1975 or H1299, or LLC cells transfected with miR-101 or scramble
To address whether miR-101 expression can suppress lung cancer cell migration, we applied wound-healing assays to the above transfected cells. When these cells became sub-confluent, a ‘wound gap’ in a cell monolayer was manually generated by scratch, followed by monitoring the ‘healing’ of wound. On the basis of cell proliferation rates, the bright-field images of the gaps were taken at 24 or 48 h after the gaps were created. As shown in Figures 5c and d, miR-101-expressing cells were much less efficient in migrating to the scratch surface than scramble cells, including 268±32 versus 876±89 μm in H1975 (P<0.01), 154±24 versus 954±92 μm in H1299 (P<0.01) or 211±24 versus 721±65 μm in LLC (P<0.05) cells, suggesting that miR-101 expression cells are not as mobile as scramble cells. In addition, as caspase-3 activation appears at the early stage of cell apoptosis, we analyzed the effect of miR-101 on caspase cleavage and found that ectopic miR-101 expression significantly increased the expression of caspase-3 active form (Figure 5e), consistent with the reported role of miR-101 in cell apoptosis.39 Notably, when H1975 and H1299 cells were transfected with miR-101 for 24 h that was followed by transfection with DNMT3a expression vectors for another 24 h, the inhibitory effects of miR-101 on lung cancer cell migration (Supplementary Figure 3a and 3b) and colony formation (Supplementary Figure 3c) were abolished by DNMT3a overexpression. Further, DNMT3a transfection also disrupted miR-101-activated caspase-3 (Supplementary Figure 3d). Thus, miR-101 suppresses lung cancer growth through DNMT3a ablation.
An increase of miR-101 leads to less tumor growth in xenograft models
To address whether miR-101 upregulation blocks lung tumor growth in vivo, LLC cells, which are syngeneic to C57BL/6 with lung metastatic potential,40 were transfected with miR-101 or scramble. At 6 h after transfection, ∼0.5 × 106 cells were subcutaneously injected into the right flank of C57BL/6 (n=6). The tumors were measured in every 4 days after injection and all mice were killed at day 28 after engraftment (Figure 6a). Although the life of miR oligos is relatively short, miR-mediated inhibition of tumorigenesis has been demonstrated.7, 41 Indeed, although tumor incidence was similar for both miR-101 and scramble (6/6, 100%), we observed that the tumor latency was different, with tumor detectable at 4 days in scramble, but at 8 days in miR-101 group after injection (Figure 6b). Tumor volume increased proportionally between both groups over 28 days, but tumors in scramble group grew faster with larger final tumor volume than that in miR-101 group. Further, the mice engrafted with miR-101-transfected cells had much lower tumor weight compared with that of scramble (551±93 versus 106±72 mg, P<0.01) (Figures 6c–e). Hematoxylin and eosin (H&E) staining of tumor sections showed that miR-101 expression remarkably impaired the tumor formation (Figure 6f). In response to miR-101 overexpression, IHC-stained tumors revealed significant difference in proliferative indices when Ki67 was used as a cellular proliferation marker (Figure 6g). By qPCR and Dotblot, significant reduction of DNMT3a expression (62±8%, P<0.05) and global DNA methylation (64±11%, P<0.05) were demonstrated between miR-101 and scramble groups (Figures 6h and i). Notably, miR-101 upregulation was verified in tumors from miR-101-transfected cells (Figure 6j). These results suggest that miR-101 re-expression promotes tumor regression via its DNA hypomethylating effects.
MiR-101 overexpression inhibits lung cancer xenograft tumor growth. (a) C57BL/6 mice (n=6 mice per group) were subcutaneously injected with 0.5 × 106 LLC cells transiently transfected with miR-101 or scramble. After implantation for 4 days, tumor volume measurement began and the experiments were terminated in 28 days. (b) Pictured is the external view of tumor-bearing C57BL/6 mice. (c) Latency of tumor formation (n=6 mice/group). Data represents mean±S.D., P<0.01. (d) Histograms represent final tumor weight from all mice (n=6 tumor/group). Bar represents mean tumor weight±S.D., P<0.01. (e) Vision analysis of tumors (n=6 tumor per group). (f) Representative images of H&E-stained xenograft tumor sections (left: magnification × 200; right: magnification × 400; n=6 tumors per group). (g) Representative images of IHC-stained xenograft tumor sections with Ki-67 antibody (left: magnification × 200; right: magnification × 400; n=6 tumors per group). (h) qPCR analysis of DNMT3a expression from xenograft tumors. The data are represented as mean±S.D., *P<0.05 (n=6 tumors per group). (i) Dotblot analysis of 5mC levels from xenograft tumors. The data are represented as mean±S.D., *P<0.05 (n=6 tumors per group). (j) qPCR analysis of miR-101 expression from xenograft tumors. The data are represented as mean±S.D., *P<0.05 (n=6 tumors per group)
Ectopic miR-101 expression inhibits LLC lung metastatic growth in mouse xenografts
Given that miR-101 overexpression suppresses cell motility in vitro (see Figures 5c and d), we hypothesized that in response to miR-101, lung cancer cell metastasis in vivo could be suppressed. To test this, LLC cells were transfected with miR-101 or scramble, and ∼0.1 × 106 cells were injected through tail vein into C57BL/6 mice. The mice were killed for dissection 18 days after cell inoculation (Figure 7a). We found that the body weight of mice inoculated with scramble cells was decreased, whereas it was slightly increased in mice engrafted with miR-101-expressing cells, an indicator of poorer prognosis in scramble than in miR-101 group (Figure 7b). Significantly higher tumor incidence (97.5% versus 44%, P<0.05) was also observed in the lungs from scramble cells when compared with that in miR-101 cells (Figures 7c and d). Further, the metastatic foci were obviously visible on the surface of lungs collected from mice inoculated with scramble cells (8.6±3.3 versus 0.7±1.1, P<0.01) (Figure 7e). H&E-stained lung tissues showed that compared with the number of pulmonary metastatic nodules found in mice injected with scramble cells, the mice injected with miR-101-expressing cells displayed fewer nodules and smaller lesion areas in the lung (Figure 7f), together, suggesting that miR-101-expressing cells have a weaker capability to metastasize into lungs. Of note, these observed anti-metastatic effects could be attributed to intrinsic difference in cell proliferation rates and migration ability, as miR-101-expressing cells exhibited a significant growth disadvantage in colony formation and much less ability in wound healing.
Effect of miR-101 on lung metastasis in C57BL/6 mouse xenograft model. (a) LLC cells were transfected with miR-101 or scramble. Tumor-bearing mice were generated by injecting 0.1 × 106 of transfected cells through tail vein into C57BL/6 mice (n=9 mice per group). Mice were killed to collect the lungs at 18 days after the cell inoculation. (b) Histogram indicates the change of body weight in tumor-bearing mice. The data are represented as mean±S.D., *P<0.05 (n=9 mice per group). (c) Images of lung from mice injected with LLC cells transfected with miR-101 or scramble showing the lung metastases. Arrows indicate focal tumor nodules on lung surfaces (n=9 mice per group). (d) Pie graphs indicate the tumor incidence of metastatic growth in lung from tumor-bearing mice, P<0.05 (n=9 mice per group). (e) The quantification of tumors from the lung of tumor-bearing mice. The data are represented as mean±S.D., P<0.01 (n=9 mice per group). (f) Representative images for H&E-stained sections of lung from c, which indicate the lung metastasis foci. Arrows indicate focal tumor nodules on lung surfaces (upper: magnification × 40; lower: magnification × 200; n=9 mice per group)
MiR-101 overexpression sensitizes lung cancer cells to decitabine treatment
Although DNMTs exhibit distinctive functions with DNA methylation maintenance of DNMT1 and de novo DNA methylation of DNMT3a or DNMT3b, it is widely accepted that DNA methylation is cooperatively controlled by all of them.9, 42 Given that decitabine specifically targets DNMT1, but not DNMT3a and DNMT3b, the repression of DNMT3a by miR-101 led to a hypothesis that there is a synergistic potential between decitabine and miR-101. To confirm this, we transfected H1975 or H1299 cells with miR-101 or scramble for 12 h, and the transfected cells were treated with 1 μM decitabine. Wound-healing assays showed that the cells treated with both miR-101 and decitabine had the least ability to migrate toward the ‘scratch’ area, when compared with the single agent (Figures 8a and b). qPCR uncovered a synergistic downregulation of DNMT3a (Figure 8c) and DNMT1 (Figure 8d), and a synergistic upregulation of CDH1 (Figure 8e). Further, Dotblot demonstrated that the global DNA methylation was synergistically decreased (Figures 8f and g). These results suggest that lung cancer patients with DNMT1 and DNMT3a overexpression may be benefited from the combination of decitabine with miR-101.
MiR-101 synergizes with decitabine to modulate DNA methylation profile and lung cancer cell growth. (a and b) Wound-healing assay for H1975 or H1299 cells treated with vehicle, miR-101 only, decitabine only, or miR-101 plus decitabine. Representative images are shown in a. Graphs are the quantification of ‘gap closure’ (b). (c) qPCR analysis for the expression of DNMT3a in H1975 or H1299 cells treated with vehicle, miR-101 only, decitabine only, or miR-101 plus decitabine. (d) qPCR analysis for the expression of DNMT1 in H1975 or H1299 cells treated with vehicle, miR-101 only, decitabine only, or miR-101 plus decitabine. (e) qPCR analysis for the expression of CDH1 in H1975 or H1299 cells treated with vehicle, miR-101 only, decitabine only, or miR-101 plus decitabine. (f and g) The genomic DNA was prepared from H1975 or H1299 cells treated with vehicle, miR-101 only, decitabine only, or miR-101 plus decitabine and subjected to Dotblot using 5mC antibody. Left: representative images are shown (f); graph is the quantification of Dotblot intensity from g. The data are represented as mean±S.D. from three independent experiments
Discussion
Despite the impressive progress made in lung cancer diagnosis and therapy, the dismal 5-year survival rate for lung cancer patients has not substantially changed. Thus, the molecular characterization of lung tumorigenesis and the identification of biomarkers for the selection of patients for targeted therapy could improve such dismal outcomes. Although DNMT3a overexpression and miR-101 silencing occur in lung cancer,7, 8, 12, 14, 30 the pathological contributions and the molecular links of these aberrations remain elusive. In this report, we identified DNMT3a as a bona fide target of miR-101 in lung cancer. We demonstrated that miR-101 restoration reduces DNMT3a expression, resulting in the global and gene-specific DNA hypomethylation. We proved that the functional interactions between miR-101 and DNA methylation determine lung cancer cell fate in vitro and in vivo via CDH1 re-expression. Although previous investigations have established the molecular linkage between miR-101 deregulation and EZH2-dependent histone modifications,29, 30, 32 our findings connect the miR-101 silencing to DNA hypermethylation, another major epigenetic code, in lung cancer cells and highlight miR-101-DNMT3a axis as a new molecular pathway for targeted therapy.
MiRs are the endogenous inhibitors of target gene expression and key regulators of most cellular processes, including cell growth and differentiation.21, 22 The miR expression patterns are found to be associated with the cancer types and stages, thereby being explored as biomarkers for cancer diagnosis and prognosis.7, 43, 44, 45, 46, 47, 48 Specifically, miR-101 is downregulated in multiple tumor types,28, 29, 30, 31, 33 and miR-101 overexpression impairs tumor cell expansion.29, 33, 49 In agreement with this, our data show that DNMT3a levels were highly elevated, while miR-101 expression was greatly reduced in human lung cancer patients and cell lines. Human and mouse lung cancer cells expressing miR-101 had a decrease of DNMT3a expression and displayed suppressed cell proliferation in vitro. When these miR-101-expressing cells were inoculated into mice, the tumor growth was remarkably attenuated and the metastasis to lung was significantly impaired, thus demonstrating the crucial contribution of miR-101 deregulation to lung cancer pathogenesis.
Although multiple pathways, including Wnt/β-catenin28 and metastasis suppressor RKIP,50 likely mediates the anti-tumor activity of miR-101, the most important molecule could be EZH2,29, 33, 49, 51 a histone-lysine N-methyltransferase that is heavily involved in cancer progression.52, 53 It is postulated that miR-101 silencing in cancers causes EZH2 overexpression that is followed by TSG inactivation through histone methylation. However, these findings are insufficient to explain why DNMTs are upregulated and the DNA methylation, including TSG promoter methylation, is increased in lung cancer. To this end, we employed qPCR for fresh-frozen tissues and cell lines, IHC staining for human lung cancer TMAs, to assess the expression of DNMT3a and miR-101. These investigations revealed that miR-101 levels were negatively correlated with DNMT3a expression, suggesting that DNMT3a and miR-101 have functional and regulatory interactions in lung tumorigenesis. In fact, our results proved that DNMT3a was a bona fide target gene of miR-101 in lung cancer cells, as miR-101 bound to DNMT3a 3′-UTR and inhibited the luciferase activity driven by DNMT3a 3′-UTR. When miR-101 was introduced into H1975 or H1299, or LLC cells, the endogenous DNMT3a expression was decreased, leading to the upregulation of CDH1 and the reduction of both global and CDH1-specific DNA methylation. These findings add a new layer to the complexity of mechanisms by which miR-101 silencing promotes lung cancer pathogenesis, and establish a role for miR-101 in regulating DNA methylation in lung cancer cells. Although it requires further demonstration, we cannot exclude the possibility that miR-101 modulates DNA methylation indirectly through EZH2, as EZH2 has been shown to physically interact with all three DNMT isoforms.54 Given the negative role of miR-101 in EZH2 expression, miR-101 loss could enhance the physical interactions between EZH2 and DNMTs, thus facilitating DNMT binding to CpG-enriched DNA regions (i.e., EZH2-target gene promoters) followed by DNA hypermethylation and TSG suppression. Regardless, our findings suggest that miR-101 inactivation enhances DNA methylation through direct suppression of DNMT3a in lung cancer cells.
Although we show that DNMT3a is a direct target of miR-101, DNMT3a is also a target of other miRs, including miR-29b and miR-30c, which are downregulated in lung cancer.7, 55 Thus, it is possible that DNMT3a-dependent DNA methylation is modulated by the cooperation of these or even other unidentified miRs, but is not the consequence of one single miR, such as miR-29b or miR-101. This could explain why overexpression of one miR, such as miR-101, induces only partial release of global and CDH1 promoter DNA methylation in lung cancer cells. Taken together, our results offer a first glimpse of a mechanistic and functional link between miR-101 dysregulation and aberrant DNA methylation in lung cancer and provide an innovative explanation for why miR-101 silencing and DNMT3a overexpression predict poor prognosis in lung cancer patients.
Materials and Methods
Cell lines and plasmids
Cell lines (H1975, H1299, A549, H1650, H520, H460, H441, H358, H69, H82 and LLC) were obtained from American Type Culture Collection. H1299, H1975, A549, H1650, H520, H460 H441, H358, H69 and H82 cells were grown in RPMI1640 containing 10% fetal bovine serum (FBS) (Life Technologies, Grand Island, NY, USA). LLC cells were cultured in DMEM supplemented with 10% FBS. MRC-5 human normal lung fibroblasts were cultured with EMEM containing 10% FBS. Luciferase plasmid containing DNMT3a 3′-UTR with miR-101 complementary site was purchased from GeneCopoeia (Rockville, MD, USA).
Lung tissue microarray
Human lung TMA slides were purchased from US Biomax Inc. (Rockville, MD, USA). These TMAs contain 45 cases of non-small cell carcinoma (NSCLC, 26 cases of squamous cell carcinoma, 18 cases of adenosquamous carcinoma, 1 case of large cell carcinoma) at different stages (18 cases of Stage I, 16 cases of Stage II, 10 cases of Stage III and 1 case of Stage IV), the adjacent tissues, and 5 normal tissues with duplicate core per cases. The IHC staining and H-score methods were described in ‘Hematoxylin and eosin and immunohistochemistry’ section.
Transfection
Targetplus Smart pool siRNAs (ON_TARGETplus siRNA, Thermo Scientific, Waltham, MA, USA) containing a mixture of four oligonucleotides with potential for human DNMT3a abrogation was used to silence DNMT3a expression. DNMT3a siRNA and miR-101, as well as their corresponding negative controls were purchased from Thermal Scientific Dharmacon RNAi Technologies (Thermo Scientific). Approximately 1 × 106 of H1975 or H1299 cells were seeded into six-well plates for overnight before transfection. A total of 100 nM of siRNA or miR-101, or the corresponding negative control was introduced into cells using Lipofectamine RNAiMAX Reagent (Life Technologies) according to the manufacturer’s instruction. DNMT3a overexpression plasmid or its empty vector was transfected into cells by Lipofectamine 2000 reagent (Life Technologies).
Reporter gene assays
Approximately 5 × 104 of H1975 or H1299 cells were seeded into 24-well plates for overnight and co-transfected with 0.1 μg of DNMT3a 3′-UTR luciferase plasmid plus miR-101 or its scramble. pRL-TK Renilla luciferase plasmid (Promega, Madison, WI, USA) was co-transfected for internal control. Firefly luciferase activity was measured 48 h after transfection by using a dual luciferase reporter assay system (Promega), according to the manufacturer’s instruction. Values were corrected to account for differences in transfection efficiency by pRL-TK Renilla luciferase activity. All luciferase data represent the average of three independent experiments.
Colony-forming assays
After transfection, about 1000 cells were subjected to the methylcellulose colony formation assays in triplicate using human methylcellulose media (R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s instructions. Colonies were scored in 10 to14 days.
Wound-healing assays
Lung cancer cells were transfected as indicated and grown to sub-confluence. The linear wound of cellular monolayer was created by scratching the sub-confluent cell monolayer using a plastic pipette tip (200 μl). Scratched cell monolayer was washed by PBS to remove debris. After incubation at 37 °C for 24 h, the migration of the cells towards the wound was photographed under light microscope. CorelDRAWX5 Software (Corel Corporation, Ottawa, ON, Canada) was used to determine the migration distance.
Hematoxylin and eosin and immunohistochemistry
Tumors and tissues collected from our animal studies were fixed in 10% neutral buffered formalin. The paraffin-embedded samples were cut to 5 μm thickness and stained with H&E. For IHC staining, slides were heated in a dry oven at 60 °C for 30 min, deparaffinized in Histo-Clear for 10 min, rehydrated through graded ethanol and rinsed in deionized water. All section slides were then microwaved in 10 mM citric acid buffer (pH 6.0) at 70% power for 10 min to unmask antigens. Endogenous peroxidase was quenched using 3% hydrogen peroxide for 20 min followed by rinsing with PBS. Nonspecific binding was blocked with 10% goat serum for 1 h, then with avidin and biotin (Vector Laboratories, Burlingame, CA, USA) for 15 min each. Primary antibodies against DNMT3a (1 : 500, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or Ki-67 (1 : 500, Abcam, Cambridge, MA) were incubated at 4 °C overnight. For the detection of primary antibodies, the Vectastain Elite ABC-peroxidase Rabbit IgG Kit (Vector Laboratories) was used. Samples were developed with 3, 3′-diaminobenzidine (Vector Laboratories), counterstained with hematoxylin and mounted. For the quantification of IHC staining, the modified H-score was used and defined as follows: no staining, no appreciable staining in normal or tumor cells; weak staining, barely detectable staining in cytoplasm and/or nucleus; moderate staining, readily appreciable brown staining distinctly marking the tumor cell cytoplasm and/or nucleus; strong staining, dark brown staining of cytoplasm and/or nucleus. The H-score was obtained using the following formula: H-score=(% cells no staining × 0)+(% cells weak staining × 1)+(% cells moderate staining × 2)+(% cells strong staining × 3). The resulting scores range from 0 (100% negative cells) to 300 (100% strong staining).
In situ hybridization
In situ hybridization (ISH) for miR-101 staining was performed on lung cancer TMA slides using miRCURY LNA microRNA ISH Optimization Kit (EXIQON, Woburn, MA, USA) according to the instruction manual v2.0. Briefly, slides were deparaffinized in xylene and ethanol, then incubated with proteinase-K at 37 °C for 10 min. After dehydration in graded ethanol, slides were hybridized with digoxigenin anti-sense miR-101 probe at 51 °C overnight, followed by stringent washing in 5, 1 and 0.2 × SSC buffer. The sections were incubated with anti-digoxigenin-AP antibody (Roche Diagnostics, Indianapolis, IN, USA) at room temperature for 1 h and developed with nitroblue tetrazolium and bromochloroindolyl phosphate (Roche Diagnostics). Stained slides were viewed and photographed with a Leica microscope mounted with a high-resolution spot camera, which is interfaced with a computer loaded with Image-Pro Plus software (Media Cybernetics, Inc., Rockville, MD, USA).
Western blotting
After the various treatments, the cells were collected into 1 × cell lysis buffer41 and the western blotting was performed with whole-cell lysates as previously described.41 Equivalent gel loading was confirmed by probing with β-actin antibody. The antibodies used were: β-actin (1 : 1000) and DNMT3a (1 : 1000) (Santa Cruz Biotechnology); EZH2 (1 : 500) and cleaved caspase-3 (1 : 500) (Cell Signaling Technology, Danvers, MA, USA).
Dotblot
The genomic DNA was purified using DNA Blood/tissue Kit (Qiagen, Gaithersburg, MD, USA) and the concentrations were accurately determined by NanoDrop. Equal amount of DNA (∼1–2 μg) from all samples was denatured in 1 × buffer (0.4 M NaOH, 10 mM EDTA) at 100 °C and mixed with an equal volume of cold 2 M ammonium acetate. After the membrane was rehydrated with 500 μl Tris-EDTA or H2O, the denatured DNA in a 50–500 μl solution was loaded onto the membrane. Next, the membrane was washed by 2 × SSC buffer, baked at 80 °C for 2 h and blocked with 5% non-fat milk for 1 h. The DNA spotted membrane was incubated with mouse anti-5mC antibody (1 : 2500, Active Motif, Carlsbad, CA, USA) and the signal was detected by HRP-conjugated secondary antibody and enhanced chemiluminescence. After development, the membrane was hybridized with 0.02% methylene blue (Sigma-Aldrich, St. Louis, MO, USA) in 0.5 M sodium acetate (pH 5.0) to stain DNA as a loading control. The Dotblot was quantified by NIH Image J Software (Bethesda, MD, USA).
Bisulfite sequencing analysis
Approximately 2 μg of genomic DNA was modified with sodium bisulfite using an EpiTect Bisulfite Kit (Qiagen). The human CDH1 promoter region (transcription start site −251 to +139) was amplified by PCR using the bisulfite-treated DNA as template. Primer sequences are:
forward: 5′-TTTTTTTTGATTTTAGGTTTTAGTGAG-3′,
reverse: 5′-ACTCCAAAAACCCATAACTAACC-3′.
PCR products were subcloned and then sequenced.
RNA isolation, cDNA preparation and qPCR
RNA was isolated using miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Reverse transcription for obtaining cDNA was performed by using SuperScript III First-Strand Synthesis System (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions. For the expression of DNMT3a and miR-101 in cell lines and fresh-frozen patient tissues, real-time qPCR was carried out by TaqMan technology (Applied Biosystems, Foster City, CA, USA). The expression of CDH1 gene was measured by SYBR Green Quantitative PCR (SYBR Green PCR Master Mix and SYBR Green RT-PCR Reagents Kit, Life Technologies). The expression of miR-101 was normalized by U44/48 (for human) or Sno202 (for mouse) levels, and the expression of DNMT3a and CDH1 were normalized by the levels of 18S. The sequences for the primers are as follows:
human CDH1: forward 5′-AGAACGCATTGCCACATACAC-3′,
human CDH1: reverse 5′-GAGGATGGTGTAAGCGATGG-3′;
mouse CDH1: forward 5′-AATGGCGGCAATGCAATCCCAAGA-3′,
mouse CDH1: reverse 5′-TGCCACAGACCGATTGTGGAGATA-3′;
human 18S: forward 5′-ACAGGATTGACAGATTGA-3′,
human 18S: reverse 5′-TATCGGAATTAACCAGACA-3′; and
mouse 18S: forward 5′-ACAGGATTGACAGATTGA-3′,
mouse 18S: reverse 5′-TATCGGAATTAACCAGACA-3′.
In vivo tumorigenicity assays
C57BL/6 mice (female, 4–6 weeks old) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). All animal experiments were performed in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals and were approved by the University of Minnesota Institutional Animal Care and Use Committee. A total of 0.5 × 106 transfected LLC cells were injected subcutaneously into the flanks of mice for xenograft tumor or 0.1 × 106 transfected LLC cells were injected intraperitoneally into the mice for metastasis. Tumor growth was monitored. Tumor diameters were measured with digital caliper and tumor volume was calculated using the formula: tumor volume (mm3)=(length × width × height × π/6).56 All tumors were excised at the same time. One-half of each tumor was frozen at −80 °C for DNA or RNA, or protein extraction, and the remainder was fixed in formalin. The lung sections were collected, and together with the tumors, fixed in 10% neutral-buffered formalin and stained with H&E (Thermo Scientific). H&E-stained sections of tumors were compared.
Lung cancer patients
The patients were diagnosed as NSCLC without smoking history at Mayo Clinic, Rochester, Mn, USA. All the cases had matched normal tissues. All the study protocols were approved by Mayo Clinic’s Institutional Review Board. All patients have been prospectively enrolled and followed for outcome research, and all human participants have written informed consent.
Statistical analysis
The quantification for gene expressions from TMA, real-time PCR, as well as other targets including Dotblot, wound-healing assay, bisulfite sequencing PCR analysis, body weight, tumor growth, tumor weight and metastatic foci were analyzed using the Student’s t-tests. The lung tumor incidence, percentage of comparison between low gene expression and high gene expression for TMA were analyzed using the χ2-test. Correlation data were performed with Pearson correlation coefficients. All statistical analyses were conducted using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Differences were considered statistically significant at P<0.05. All P-values were two-sided.
Abbreviations
- DNMT:
-
DNA methyltransferase
- TSG:
-
tumor suppressor gene
- miR:
-
microRNA
- CDH1:
-
cadherin 1
- IHC:
-
immunohistochemistry
- TMA:
-
tissue microarray
- 5mC:
-
5-methylcytosine
- NSCLC:
-
non-small cell lung cancer
- siRNA:
-
small interfering RNA
- LLC:
-
Lewis lung carcinoma
- H&E:
-
hematoxylin and eosin
- ISH:
-
in situ hybridization
References
Choong NW, Ma PC, Salgia R . Therapeutic targeting of receptor tyrosine kinases in lung cancer. Expert Opin Ther Targets 2005; 9: 533–559.
Jafri NF, Ma PC, Maulik G, Salgia R . Mechanisms of metastasis as related to receptor tyrosine kinases in small-cell lung cancer. J Environ Pathol Toxicol Oncol 2003; 22: 147–165.
Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ et al. Cancer statistics 2007 CA Cancer J Clin 2007; 57: 43–66.
Jones PA, Laird PW . Cancer epigenetics comes of age. Nat Genet 1999; 21: 163–167.
Baylin SB, Herman JG . DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 2000; 16: 168–174.
Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y et al. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 2001; 97: 1172–1179.
Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA 2007; 104: 15805–15810.
Kase S, Sugio K, Yamazaki K, Okamoto T, Yano T, Sugimachi K et al. Expression of E-cadherin and beta-catenin in human non-small cell lung cancer and the clinical significance. Clin Cancer Res 2000; 6: 4789–4796.
Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Gene Cells 2006; 11: 805–814.
Chen T, Ueda Y, Dodge JE, Wang Z, Li E . Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 2003; 23: 5594–5605.
Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA et al. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene 2005; 24: 1625–1633.
Fabbri M, Iliopoulos D, Trapasso F, Aqeilan RI, Cimmino A, Zanesi N et al. WWOX gene restoration prevents lung cancer growth in vitro and in vivo. Proc Natl Acad Sci USA 2005; 102: 15611–15616.
Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E et al. MicroRNA -29b induces global DNA hypomethylation and tumor suppressor gene re-expression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009; 113: 6411–6418.
Sandoval J, Mendez-Gonzalez J, Nadal E, Chen G, Carmona FJ, Sayols S et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol 2013; 31: 4140–4147.
Toyooka S, Matsuo K, Gazdar AF . DNA methylation in lung cancer. N Engl J Med 2008; 358: 2513.
Zhao Z, Wu Q, Cheng J, Qiu X, Zhang J, Fan H et al. Depletion of DNMT3A suppressed cell proliferation and restored PTEN in hepatocellular carcinoma cell. J Biomed Biotechnol 2010: 737535.
Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005; 37: 766–770.
Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 2005; 120: 21–24.
Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM . MicroRNA expression and function in cancer. Trends Mol Med 2006; 12: 580–587.
Pillai RS, Artus CG, Filipowicz W . Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525 2004.
Pillai R. S., Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 2005; 309: 1573–1576.
Pillai RS, Bhattacharyya SN, Filipowicz W . Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 2007; 17: 118–126.
Calin GA, Croce CM . MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857–866.
Peng Y, Dai Y, Hitchcock C, Yang X, Kassis ES, Liu L et al. Insulin growth factor signaling is regulated by microRNA-486, an underexpressed microRNA in lung cancer. Proc Natl Acad Sci USA 2013; 110: 15043–15048.
Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010; 18: 282–293.
Braconi C, Huang N, Patel T . MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010; 51: 881–890.
Duursma AM, Kedde M, Schrier M, le Sage C, Agami R . miR-148 targets human DNMT3b protein coding region. RNA 2008; 14: 872–877.
Strillacci A, Valerii MC, Sansone P, Caggiano C, Sgromo A, Vittori L et al. Loss of miR-101 expression promotes Wnt/beta-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol 2013; 229: 379–389.
Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008; 322: 1695–1699.
Thu KL, Chari R, Lockwood WW, Lam S, Lam WL . miR-101 DNA copy loss is a prominent subtype specific event in lung cancer. J Thorac Oncol 2011; 6: 1594–1598.
Schee K, Boye K, Abrahamsen TW, Fodstad O, Flatmark K . Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer 2012; 12: 505.
Cho HM, Jeon HS, Lee SY, Jeong KJ, Park SY, Lee HY et al. microRNA-101 inhibits lung cancer invasion through the regulation of enhancer of zeste homolog 2. Exp Ther Med 2011; 2: 963–967.
Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res 2009; 69: 2623–2629.
Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466: 1129–1133.
Ceteci F, Ceteci S, Karreman C, Kramer BW, Asan E, Götz R et al. Disruption of tumor cell adhesion promotes angiogenic switch and progression to micrometastasis in RAF-driven murine lung cancer. Cancer Cell 2007; 12: 145–159.
Sheikhnejad R, Zohri M, Shadmehr MB, Rahmani-Khalili M, Doozande N, Farsad Z et al. Detection of aberrant methylation of 10 genes in genomic DNA of lung tumors. Ann Oncol 2013; 24: 2705–2706.
Sulzer MA, Leers MP, van Noord JA, Bollen EC, Theunissen PH . Reduced E-cadherin expression is associated with increased lymph node metastasis and unfavorable prognosis in non-small cell lung cancer. Am J Respir Crit Care Med 1998; 157: 1319–1323.
Liu D, Huang C, Kameyama K, Hayashi E, Yamauchi A, Kobayashi S et al. E-cadherin expression associated with differentiation and prognosis in patients with non-small cell lung cancer. Ann Thorac Surg 2001; 71: 949–954.
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res 2009; 69: 1135–1142.
Bacher U, Haferlach C, Schnittger S, Kohlmann A, Kern W, Haferlach T et al. Mutations of the TET2 and CBL genes: novel molecular markers in myeloid malignancies. Ann Hematol 2010; 89: 643–652.
Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell 2010; 17: 333–347.
Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet 2003; 33: 61–65.
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006; 9: 189–198.
Navarro A, Diaz T, Gallardo E, Viñolas N, Marrades RM, Gel B et al. Prognostic implications of miR-16 expression levels in resected non-small-cell lung cancer. J Surg Oncol 2011; 103: 411–415.
Hennessey PT, Sanford T, Choudhary A, Mydlarz WW, Brown D, Adai AT et al. Serum microRNA biomarkers for detection of non-small cell lung cancer. PLoS One 2012; 7: e32307.
Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol 2011; 4: 575–586.
Tang D, Shen Y, Wang M, Yang R, Wang Z, Sui A et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev 2013; 22: 540–548.
Leidinger P, Keller A, Meese E . MicroRNAs - important molecules in lung cancer research. Front Genet 2011; 2: 104.
Banerjee R, Mani RS, Russo N, Scanlon CS, Tsodikov A, Jing X et al. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene 2011; 30: 4339–4349.
Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res 2012; 72: 3091–3104.
Kottakis F, Polytarchou C, Foltopoulou P, Sanidas I, Kampranis SC, Tsichlis PN et al. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell 2011; 43: 285–298.
Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629.
Takawa M, Masuda K, Kunizaki M, Daigo Y, Takagi K, Iwai Y et al. Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer Sci 2011; 102: 1298–1305.
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439: 871–874.
Xia Y, Chen Q, Zhong Z, Xu C, Wu C, Liu B et al. Down-regulation of miR-30c promotes the invasion of non-small cell lung cancer by targeting MTA1. Cell Physiol Biochem 2013; 32: 476–485.
Tomayko MM, Reynolds CP . Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol 1989; 24: 148–154.
Acknowledgements
This work was supported partially by the Hormel Foundation (S Liu) and National Cancer Institute (Bethesda, MD) grants R01CA149623 and R21CA155915 (S Liu).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Baehrecke
Author contributions
SJL designed the research; FY, NS, JXP, DX and BD performed research; FY, PY, JRM and SJL analyzed data and wrote the paper; PY and JRM provided the patient samples; SJL conceived the idea and supervised the whole project. All authors discussed the results and commented on the manuscript.
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Yan, F., Shen, N., Pang, J. et al. Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell Death Dis 5, e1413 (2014). https://doi.org/10.1038/cddis.2014.380
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.380
This article is cited by
-
DNMT3L inhibits hepatocellular carcinoma progression through DNA methylation of CDO1: insights from big data to basic research
Journal of Translational Medicine (2024)
-
Bridging tissue repair and epithelial carcinogenesis: epigenetic memory and field cancerization
Cell Death & Differentiation (2024)
-
Focused screening reveals functional effects of microRNAs differentially expressed in colorectal cancer
BMC Cancer (2019)
-
Regulation of PTEN expression by noncoding RNAs
Journal of Experimental & Clinical Cancer Research (2018)
-
A vicious loop of fatty acid-binding protein 4 and DNA methyltransferase 1 promotes acute myeloid leukemia and acts as a therapeutic target
Leukemia (2018)