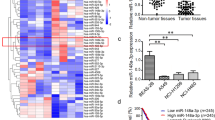
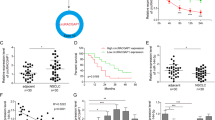
Abstract
Upregulation of RNA polymerase I (Pol I) transcription and the overexpression of Pol I transcriptional machinery are crucial molecular alterations favoring malignant transformation. However, the causal molecular mechanism(s) of this aberration remain largely unknown. Here, we found that Pol I transcription and its core machinery are upregulated in lung adenocarcinoma (LUAD). We show that the loss of miRNAs (miR)-330-5p and miR-1270 expression contributes to the upregulation of Pol I transcription in LUAD. Constitutive overexpression of these miRs in LUAD cell lines suppressed the expression of core components of Pol I transcription, and reduced global ribosomal RNA synthesis. Importantly, miR-330-5p/miR-1270-mediated repression of Pol I transcription exerted multiple tumor suppressive functions including reduced proliferation, cell cycle arrest, enhanced apoptosis, reduced migration, increased drug sensitivity, and reduced tumor burden in a mouse xenograft model. Mechanistically, the downregulation of miR-330-5p and miR-1270 is regulated by Pol I subunit-derived circular RNA circ_0055467 and DNA hypermethylation, respectively. This study uncovers a novel miR-330-5p/miR-1270 mediated post-transcriptional regulation of Pol I transcription, and establish tumor suppressor properties of these miRs in LUAD. Ultimately, our findings provide a rationale for the therapeutic targeting of Pol I transcriptional machinery for LUAD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data generated in this study are available within the article and its supplementary data files. TCGA gene expression data for LUAD is available from UCSC Xena browser. The CpG island methylation track (cg19204924) of LUAD or normal is available from TCGA Wanderer database (http://maplab.imppc.org/wanderer/).
References
Goodfellow SJ, Zomerdijk JC. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell Biochem. 2013;61:211–36.
Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–92.
White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78.
Babaian A, Rothe K, Girodat D, Minia I, Djondovic S, Milek M, et al. Loss of m(1)acp(3)Psi ribosomal RNA modification is a major feature of cancer. Cell Rep. 2020;31:107611.
Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65.
Rossetti S, Wierzbicki AJ, Sacchi N. Mammary epithelial morphogenesis and early breast cancer. Evidence of involvement of basal components of the RNA Polymerase I transcription machinery. Cell Cycle. 2016;15:2515–26.
Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–702.
Bell SP, Learned RM, Jantzen HM, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 1988;241:1192–7.
Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JC. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J. 2001;20:1373–82.
Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006;25:6384–91.
Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell. 2001;8:1063–73.
Zhao J, Yuan X, Frodin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell. 2003;11:405–13.
Lin CY, Navarro S, Reddy S, Comai L. CK2-mediated stimulation of Pol I transcription by stabilization of UBF-SL1 interaction. Nucleic Acids Res. 2006;34:4752–66.
Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharm Toxicol. 2010;50:131–56.
Sharifi S, Bierhoff H. Regulation of RNA polymerase I transcription in development, disease, and aging. Annu Rev Biochem. 2018;87:51–73.
Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–60.
Palmero EI, de Campos SG, Campos M, de Souza NC, Guerreiro ID, Carvalho AL, et al. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol. 2011;34:363–70.
Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–90.
Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–31.
Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900–D8.
L McInnes, J Healy, N Saul, L Großberger. UMAP: Uniform manifold approximation and projection. J Open Source Softw. 2018. https://doi.org/10.21105/joss.00861.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7.
Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13:e0206239.
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.
Diez-Villanueva A, Mallona I, Peinado MA. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin. 2015;8:22.
Naidu S, Shi L, Magee P, Middleton JD, Lagana A, Sahoo S, et al. PDGFR-modulated miR-23b cluster and miR-125a-5p suppress lung tumorigenesis by targeting multiple components of KRAS and NF-kB pathways. Sci Rep. 2017;7:15441.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.
Gupta RK, Kuznicki J. Biological and medical importance of cellular heterogeneity deciphered by single-cell RNA sequencing. Cells 2020;9:1751.
Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–8.
Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–90.
Honda R, Lowe ED, Dubinina E, Skamnaki V, Cook A, Brown NR, et al. The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles. EMBO J. 2005;24:452–63.
Kumazawa T, Nishimura K, Katagiri N, Hashimoto S, Hayashi Y, Kimura K. Gradual reduction in rRNA transcription triggers p53 acetylation and apoptosis via MYBBP1A. Sci Rep. 2015;5:10854.
Schuler M, Bossy-Wetzel E, Goldstein JC, Fitzgerald P, Green DR. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J Biol Chem. 2000;275:7337–42.
Prakash V, Carson BB, Feenstra JM, Dass RA, Sekyrova P, Hoshino A, et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat Commun. 2019;10:2110.
Drapela S, Bouchal J, Jolly MK, Culig Z, Soucek K. ZEB1: A critical regulator of cell plasticity, DNA damage response, and therapy resistance. Front Mol Biosci. 2020;7:36.
Suzuki H, Maruyama R, Yamamoto E, Kai M. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. 2013;4:258.
Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585:2087–99.
Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26:29.
Penzo M, Montanaro L, Trere D, Derenzini M. The ribosome biogenesis-cancer connection. Cells. 2019;8:55.
Ferreira R, Schneekloth JS, Jr., Panov KI, Hannan KM, Hannan RD. Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cells. 2020;9:266.
Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, et al. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell. 2005;19:77–87.
Low JY, Sirajuddin P, Moubarek M, Agarwal S, Rege A, Guner G, et al. Effective targeting of RNA polymerase I in treatment-resistant prostate cancer. Prostate 2019;79:1837–51.
Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–29.
Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219–35.
Zhang M, Han Y, Zheng Y, Zhang Y, Zhao X, Gao Z, et al. ZEB1-activated LINC01123 accelerates the malignancy in lung adenocarcinoma through NOTCH signaling pathway. Cell Death Dis. 2020;11:981.
Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–11.
Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–42.
Chi Y, Zheng W, Bao G, Wu L, He X, Gan R, et al. Circular RNA circ_103820 suppresses lung cancer tumorigenesis by sponging miR-200b-3p to release LATS2 and SOCS6. Cell Death Dis. 2021;12:185.
Acknowledgements
We are grateful to IIT Ropar for funding. We thank the animal facility at IISER, Mohali, Punjab, India for in-vivo studies. We thank Dr. Juhi Tayal and Dr. Anurag Mehta, Biorepository facility, RGCIRC, New Delhi, India, for providing tumor samples. We heartfully thank Prof. Galande IISER Pune, for H23 cells and Dr. Harshan, CCMB Hyderabad, for ZEB1 expression vector. Special thanks to Arpita Karmakar and Deepika for their help during revision work, and Sheetal Yadav for manuscript editing.
Funding
Seed grant—IIT Ropar granted to Dr. Srivatsava Naidu.
Author information
Authors and Affiliations
Contributions
S.N. conceived and designed the study, interpreted the data, drafted and revised the manuscript. S.S. acquired data, handled animal work and played role in interpreting the results, drafted the manuscript. E.G. acquired data and prepared figures. S.S.S. acquired data, played a role in computational analysis. S.C. and A.C. contributed in animal work and project management. S.K., G.A. contributed in computational analysis. All authors approved the final version, agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Mice work protocols were approved (IISERM/SAFE/PRT/2020/005) by the institutional animal ethics committee, Indian Institute of Science Education and Research (IISER) Mohali, as per the standards of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), and comply with the ARRIVE guidelines. Frozen specimens of LUAD or NAT were obtained from Rajiv Gandhi Cancer Institute and Research Centre (RGCI&RC), New Delhi, India, in accordance with the guidelines of the institutional review board (Approval number: RGCIRC/IRB/343/2019) and the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saproo, S., Sarkar, S.S., Gupta, E. et al. MiR-330-5p and miR-1270 target essential components of RNA polymerase I transcription and exhibit a novel tumor suppressor role in lung adenocarcinoma. Cancer Gene Ther 30, 288–301 (2023). https://doi.org/10.1038/s41417-022-00544-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-022-00544-4