Abstract
Lung adenocarcinoma (LUAD) characterized by high metastasis and mortality is the leading subtype of non-small cell lung cancer. Evidence shows that some microRNAs (miRNAs) may act as oncogenes or tumor suppressor genes, leading to malignant tumor occurrence and progression. To better understand the molecular mechanism associated with miRNA methylation in LUAD progression and clinical outcomes, we investigated the correlation between miR-148a-3p methylation and the clinical features of LUAD. In the LUAD cell lines and tumor tissues from patients, miR-148a-3p was found to be significantly downregulated, while the methylation of miR-148a-3p promoter was notably increased. Importantly, miR-148a-3p hypermethylation was closely associated with lymph node metastasis. We demonstrated that mitogen-activated protein (MAP) kinase kinase kinase 9 (MAP3K9) was the target of miR-148a-3p and that MAP3K9 levels were significantly increased in both LUAD cell lines and clinical tumor tissues. In A549 and NCI-H1299 cells, overexpression of miR-148a-3p or silencing MAP3K9 significantly inhibited cell growth, migration, invasion and cytoskeleton reorganization accompanied by suppressing the epithelial-mesenchymal transition. In a nude mouse xenograft assay we found that tumor growth was effectively inhibited by miR-148a-3p overexpression. Taken together, the promoter methylation-associated decrease in miR-148a-3p could lead to lung cancer metastasis by targeting MAP3K9. This study suggests that miR-148a-3p and MAP3K9 may act as novel therapeutic targets for the treatment of LUAD and have potential clinical applications.
Similar content being viewed by others
Introduction
Non-small cell lung cancer (NSCLC) accounts for ~85% of all kinds of lung cancers [1]. Lung adenocarcinoma (LUAD), characterized by high metastasis and mortality, is the leading subtype of NSCLC [2]. The low survival rate for NSCLC patients is mainly due to distant metastasis that accounts for 90% of cancer-related deaths [2]. Although many therapeutic strategies have been applied, the prognosis of NSCLC remains unsatisfactory. Lack of appropriate biomarkers and accurate therapeutic targets and toxicity of chemotherapy drugs lead to poor curative efficacy in lung cancer [3]. Therefore, it is of great significance to explore the molecular mechanism of LUAD occurrence and progression and discover promising therapeutic targets to improve its prognosis.
MicroRNAs (miRNAs) are small (20–24 nucleotides) endogenous non-coding RNAs that regulate vital biological processes, such as cell cycling, growth, apoptosis, and the regulation of gene expression [4,5,6]. miRNAs function in RNA silencing mainly via complementary binding to the 3′-untranslated region (3′-UTR) of its target mRNA molecules [4]. Lung cancer occurs through a multi-step process, which is related to either the activation of oncogenes or silencing of tumor suppressors [7]. Some miRNAs may be considered oncogenes or tumor suppressor genes, leading to malignant tumor occurrence or suppression. For instance, downregulation of miR-148a-3p mediates tumorigenesis and progression in several types of cancer. It also suppresses cell proliferation and migration by targeting erb-b2 receptor tyrosine kinase 3 in colorectal cancer [8]. In addition, it suppresses NSCLC metastasis both in vitro and in vivo [9].
Mitogen-activated protein kinase (MAPK) kinase kinase kinase 9 (MAP3K9), also named MLK1, regulates the growth, differentiation, migration, and apoptosis of cells [10]. Recently, it was reported that inhibition of MAP3K9 suppresses cell necroptosis in pancreatic cancer [11]. Moreover, miR-34a functions as a tumor suppressor by inhibiting MAP3K9 expression in neuroblastoma [12]. Although these reports have suggested that MAP3K9 functions as an oncogene in tumors, its underlying molecular mechanism remains unclear. Therefore, it is crucial to investigate the tumor-target regulation of MAP3K9 and reveal its potential clinical applications.
Epigenetic regulation is an important driving factor in the early stages of tumorigenesis. DNA methylation is one of the key epigenetic mechanisms that affects gene expression in mammals [13]. More importantly, aberrant DNA methylation significantly affects epigenetic regulation in cancer, resulting in aberrant gene expression [14]. Consequently, silencing tumor suppressor miRNAs via promoter hypermethylation is becoming a significant marker of tumors. Whether the methylation of the miR-148a-3p promoter participates in LUAD metastasis has not yet been studied. Therefore, in the current study, we evaluated whether epigenetic modification of miR-148a-3p is linked to promoter methylation and explored the molecular mechanisms of miR-148a-3p and MAP3K9 in LUAD in vitro and in vivo.
Materials and methods
Materials
The experimental mice were obtained from Beijing HFK Bioscience (Beijing, China). 293 T, A549, NCI-H1299, NCI-H460, and BEAS-2B cell lines were from the Cell Bank of Chinese Academy of Sciences. Phosphate buffer solution (PBS), DMEM, fetal bovine serum (FBS), TRIzol, Pierce BCA protein assay kit, Lipofectamine 3000 and SuperSignal chemiluminescent substrate were from Thermo Fisher (Waltham, MA, USA). Penicillin and streptomycin were from Sigma-Aldrich (St. Louis, MO, USA). SYBR Green qPCR kit and Evo M-MLV RT kit were bought from Accurate Biotechnology (Changsha, China). TUNEL assay kit was purchased from Servicebio (Wuhan, China). Cell counting kit-8 (CCK8) and 12-hydroxylauric acid were from Yeasen Biotech (Shanghai, China). Transwell inserts (pore size 8.0 μm) were bought from Corning (Corning, NY, USA). miR-148a-3p mimic and its scramble control (miR-NC), siRNA targeting MAP3K9 or DNMT1 (si-MAP3K9, si-DNMT1) and a scramble control (si-NC) were from Genepharma (Shanghai, China). Dual luciferase reporter assay kit and pGL3 vector were from Promega (Madison, WI, USA). The Entranster-in vivo transfection reagent was bought from Engreen Biosystem (Beijing, China). The primary antibodies used in this study were anti-GAPDH (Sigma-Aldrich, St. Louis, MO, USA), anti-MAP3K9 (Affinity Biosciences, Beijing, China), anti-Vimentin, -Snail, -E-cadherin and -N-cadherin (ImmunoWay Biotechnology, Plano, TX, USA). The HRP-conjugated secondary antibodies were from Proteintech (Wuhan, China).
Patients and clinical samples
We obtained 56 pairs of tumor and tumor-adjacent normal tissues from patients with LUAD from the Affiliated Cancer Hospital of Guangzhou Medical University. We conducted this study with the approval of the Ethics Committee of the Affiliated Cancer Hospital of Guangzhou Medical University (202108004). Written informed consent was obtained from each participant prior to the study.
miRNA microarray assay
Four pairs of tumor and adjacent normal tissues from patients with LUAD were used for miRNA microarray (Sinotech Genomics). In brief, total RNA of each sample was extracted using miRNeasy mini kit (Qiagen). Paired-end libraries were synthesized by QIAseq miRNA library kit (Qiagen) and then sequenced on Illumina HiSeq X ten platform. miRNA expression analysis was performed using DESeq software. miRNAs from tumor and adjacent tissue with log2(FC) value > 1 and <0.05 were considered as significantly modulated.
Cell culture and transfection
LUAD (A549, NCI-H1299, and NCI-H460) and BEAS-2B cells were cultured in the Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were seeded into 12-well plates and transfected using Lipofectamine 3000 at ~70% confluence.
Transwell migration and invasion assays
Transwell migration and invasion assays were performed, according to the previously described protocols [15]. In brief, the upper chambers were coated with Matrigel for the invasion assays and without Matrigel for the migration assays. The invading cells that adhered to the lower surface of the membrane were stained and counted.
Western blotting assay
Samples with equal amounts of total protein were separated using a universal approach [15]. The primary antibodies used were as follows: anti-E-cadherin (1:1000, 135 kDa), anti-N-cadherin (1:1000, 140 kDa), anti-Snail (1:1000, 29 kDa), anti-MAP3K9 (1:1000, 121 kDa), anti-vimentin (1:1000, 57 kDa), and anti-GAPDH (1:1000, 37 kDa). The SuperSignal chemiluminescent substrate was used to visualize the protein of interest after the horseradish peroxidase-conjugated secondary antibodies were bound to the primary antibodies.
RT-qPCR assay
RNA extraction and RT-qPCR assay were performed according to the previously described protocols [15]. Primers used for RT-qPCR were shown in Table 1. Differences among target expression were quantitatively analyzed.
Methylation-specific PCR (MSP)
The MSP assay was carried out according to the previously described protocol [16] and the primers of methylated (M) or unmethylated (U) miR‑148a-3p promoter were shown in Table 2.
Colony formation assay and cell counting kit-8 (CCK8) assay
The colony formation assay was conducted to detect the ability of colony growth according to the previous report [17]. And CCK8 assay was carried out using a universal approach [17]. The cell viability ratio was calculated as
5-Aza-deoxycytidine (5-Aza) treatment
Briefly, cells were seeded in six-well plates 24 h before 5-Aza (5 μmol/L) treatment. RNA was extracted 48 h after 5-Aza treatment to examine miR-148a-3p and MAP3K9 expression.
Luciferase reporter assay
The luciferase reporter assay was conducted as described previously [18]. The DNA fragments containing the wild-type (WT) or mutant (MT) 3ʹ-UTR of MAP3K9 were chemically synthesized and inserted into the pGL3 vector to generate WT MAP3K9-3ʹ-UTR and MT MAP3K9-3ʹ-UTR reporters.
Tumor xenografts
All the experimental procedures were previously approved and supervised by the Animal Experiment Ethics Committee of Guangzhou Medical University (GY2021-028, Guangzhou, China) according to ARRIVE guidelines. Six-week-old male athymic nude mice were used for xenograft studies. A total of 2 × 106 NCI-H1299 or NCI-H1299 cells stably overexpressing miR‑148a-3p (NCI-H1299miR-148a-3p) were suspended in 200 μL phosphate-buffered saline (PBS) and subcutaneously injected into the right flank regions of mice to generate NCI-H1299 and NCI-H1299miR-148a-3p tumor-bearing mice, respectively. After 14 d, the NCI-H1299 tumor-bearing mice were randomly divided into three groups for additional treatment: (1) saline (n = 5); (2) free miR-148a-3p (treated with miR-148a-3p dissolved in saline, n = 5, 20 μg miR-148a-3p per mouse); (3) encapsulated miR-148a-3p (treated with miR-148a-3p mixed with Entranster-in vivo transfection reagent, n = 5, 20 μg miR-148a-3p per mouse). The NCI-H1299 tumor-bearing mice were injected intravenously via the tail vein every 3 days for 2 weeks. Meanwhile, NCI-H1299miR-148a-3p tumor-bearing mice were not treated. The NCI-H1299 and NCI-H1299miR-148a-3p tumor-bearing mice were euthanized and analyzed at the end of the experiment. Tumor size and body weight were measured daily throughout the experiments. Tumor volume was calculated using the formula: (width2 × length)/2. After sacrifice, the tumors, livers, spleens, kidneys, hearts, and lungs of the mice were collected and stained with haematoxylin and eosin (H&E).
Terminal deoxyribonucleotidyl transferase-mediated dUTP nick-end labelling (TUNEL) assay
The sections of tumors were labelled with the TUNEL assay kit and examined under a Carl Zeiss LSM 880 laser confocal microscope (Oberkochen, Germany).
Statistical analysis
The STATISTICA program (StatSoft) was used for the analysis of the data and all the data were expressed as mean ± standard deviations (SD). Multiple comparisons were assessed by One-way ANOVA with Tukey’s post hoc test. Correlation was calculated using the ReglinP function and inverted Student’s t test. P <0.05 was considered statistically significant.
Results
High levels of miR‑148a-3p indicate better prognosis of patients with LUAD
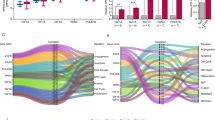
miRNA expression profiling was performed for tumor samples and matched adjacent lung samples using a microRNA microarray assay. As shown in Fig. 1a, miR-148a-3p levels were lower in LUAD tissues than in the surrounding normal lung tissues. We used reverse transcription-quantitative polymerase chain reaction (RT-qPCR) to determine miR-148a-3p levels in lung cancer and paired normal samples. The results indicated a significant downregulation of miR-148a-3p in the tumor samples, but not in the matched adjacent lung samples (Fig. 1b). The miR-148a-3p levels were also analyzed in various LUAD (A549, NCI-H1299, and NCI-H460) and non-tumorigenic lung (BEAS-2B) cell lines. As predicted, miR-148a-3p levels were significantly decreased in lung cancer cells than in BEAS-2B cells (Fig. 1c). The Cancer Genome Atlas cohort database (Oncolnc, http://www.oncolnc.org) was used to determine the correlation between miR-148a-3p levels and the overall survival time of patients with lung cancer. According to the results, lower miR-148a-3p levels were related to a dramatically worse overall survival rate in patients with lung cancer (Fig. 1d).
a Hierarchical clustering analysis of the differential expressed miRNAs on microarray assay (n = 4). b The miR-148a-3p mRNA levels were detected in lung adenocarcinoma and adjacent non-tumor tissues (n = 56). c The miR-148a-3p mRNA levels were measured in BEAS-2B, A549, NCI-H1299 and NCI-H460 cells (n = 3). d The overall survival of lung cancer patients with low and high miR-148a-3p levels (n = 245). **P < 0.01 and ***P < 0.001.
DNA methylation epigenetically regulates miR-148a-3p expression
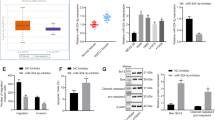
Since epigenetic silencing may contribute to the inactivation of tumor suppressors, the correlation between miR-148a-3p methylation and clinical features of LUAD was further explored. From clinical samples of 56 cases patients, we identified that DNA methylation in the miR-148a-3p promoter region occurred frequently in the tumor tissues (30/56), while the adjacent normal tissues were all unmethylated (0/56). Based on the methylation status of the miR-148a-3p promoter, we further divided the 56 clinical cases into two groups: high-methylation (30/56, 53.6%) and low-methylation (26/56, 46.4%) groups. miR-148a-3p expression levels were significantly upregulated in the low-methylation group of patients with LUAD (Fig. 2a). According to the clinicopathologic characteristics of 56 patients with LUAD, lymph node metastasis was observed in 23 patients, but not in others. The promoter methylation rates of miR-148a-3p in the lymph node metastasis and no lymph node metastasis subgroups were 69.6% and 42.4%, respectively, indicating an obvious difference (P = 0.0451). Meanwhile, methylation of the miR-148a-3p promoter was not significantly correlated with age, sex, or pathological differentiation (Table 3). Consequently, the level of miR-148a-3p methylation was strongly related to lymph node metastasis.
a The methylation status of miR-148a-3p promoter was detected in tumor samples (n = 56). b MSP detection for methylated or demethylated miR-148a-3p promoter in LUAD cells and BEAS-2B cells (n = 3). c The miR-148a-3p mRNA levels in LUAD cells and BEAS-2B cells (treated with or without 5-Aza for 24 h) were measured (n = 3). d The DNMT1 mRNA levels were measured in LUAD cells (n = 3). **P < 0.01 and ***P < 0.001.
High methylation levels of miR-148a-3p were further confirmed in LUAD cell lines, such as A549, NCI-H1299, and NCI-H460 cells, while miR-148a-3p found to be unmethylated in BEAS-2B cells (Fig. 2b). Interestingly, miR-148a-3p levels were dramatically increased after treatment with 5-Azacytidine (5-Aza) in LUAD cells, but remained unchanged in BEAS-2B cells (Fig. 2c). Compared with BEAS-2B cells, the mRNA levels of DNA methyltransferase 1 (DNMT1) were elevated in LUAD cells (Fig. 2d). Consistently, knockdown of DNMT1 by siRNA increased the expressions of miR-148a-3p in LUAD cell lines (Supplementary Fig. S1). This suggests that epigenetic regulators, such as DNMT1, can affect miR-148a-3p levels, and that methylation of the miR-148a-3p gene promoter might be an important factor in the progression of LUAD.
Mutations in the K-Ras oncogene and p53 tumor suppressor gene are among the most common genetic alterations in lung cancer that have great clinical significance with both predictive and prognostic values. Both A549 and H460 cells contained K-Ras mutations, whereas H1299 cells harbored wild-type K-Ras. Meanwhile, A549 and H460 cells express wild-type p53, while H1299 cells were p53-deficient. Therefore, A459 and H1299 cells were selected as two types of LUAD cellular models based on genetic diversity and were used in the rest of the experiments.
miR‑148a-3p targets MAP3K9
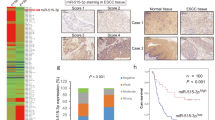
The mirDB database (http://www.mirdb.org/) was used to search for the target genes of miR-148a-3p, and MAP3K9 was identified (Fig. 3a). To validate the interaction between MAP3K9 and miR-148a-3p in LUAD cells, a luciferase reporter assay was performed. The putative wild-type and mutated 3′-UTR of the MAP3K9 gene were inserted into the luciferase reporter vector, followed by transfection of miR-148a-3p mimic into these reporter vectors. In A549 and NCI-H1229 cells transfected with the MAP3K9 3′-UTR wild-type construct, which contained the miR-148a-3p-binding site, the luciferase reporter activity was significantly decreased compared to the control mimic groups (Fig. 3b). In contrast, miR-148a-3p overexpression did not affect the luciferase activity of the mutant reporter vector (Fig. 3b). Our results also demonstrated that MAP3K9 mRNA levels were significantly downregulated after A549 and NCI-H1299 cells were treated with the miR-148a-3p mimic (Fig. 3c). Conversely, MAP3K9 mRNA levels were significantly upregulated after A549 and NCI-H1299 cells were treated with the miR-148a-3p inhibitor (Supplementary Fig. S2). Moreover, the protein levels of MAP3K9 in LUAD cells (A549 and NCI-H1299 cells) were also notably downregulated by transfection with the miR-148a-3p mimic (Fig. 3d). These results indicated that miR‑148a-3p decreased MAP3K9 expression levels in NSCLC cells by binding to the 3′-UTR of MAP3K9 mRNA. Consistent with the cellular tests, we found that the mRNA levels of MAP3K9 in the tumor tissues were also significantly upregulated compared to those in the matched normal tissues (Fig. 3e). Furthermore, patients with high MAP3K9 levels exhibited low miR-148a-3p levels. Pearson correlation analysis also demonstrated a clear negative relationship between miR-148a-3p levels and MAP3K9 mRNA levels (r2 = 0.1145, P = 0.0014) (Fig. 3f). In addition, treatment with 5-Aza inhibited MAP3K9 expression in lung cancer cells (Fig. 3g). Consistently, knockdown of DNMT1 by siRNA suppressed the mRNA and protein expression levels of MAP3K9 in A549 and H1299 cells (Supplementary Fig. S3). Taken together, these results support that miR-148-3p suppresses MAP3K9 expression in LUAD by binding to its 3′-UTR and downregulation of miR-148a-3p by DNMT1 via DNA methylation might contribute to the metastasis of LUAD through MAP3K9.
a The target genes of miR-148a-3p. b Putative miR-148a-3p binding sequence in 3′-UTR of MAP3K9 and its mutant used in the luciferase reporter assay that performed in A549 and NCI-H1299 cells (n = 3). The mRNA (c) and protein (d) levels of MAP3K9 were measured in miR-148a-3p mimic-transfected A549 and NCI-H1299 cells, respectively (n = 3). e The MAP3K9 mRNA levels were measured in lung adenocarcinoma and adjacent non-tumor tissues (n = 56). f The relationship of miR-148a-3p and MAP3K9 in LUAD tissues (n = 56). g MAP3K9 protein levels in A549 and NCI-H1299 cells treated with 5-Aza, respectively. *P < 0.05, **P < 0.01 and ***P < 0.001.
miR-148a-3p mimic or si-MAP3K9 inhibits the proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) of lung cancer cells
We determined the role of miR-148a-3p mimic or si-MAP3K9 in the growth of A549 and NCI-H1299 cells. First, cell proliferation was inhibited effectively in a time-dependent manner after treatment with miR-148a-3p mimic or si-MAP3K9 (Fig. 4a and b). Moreover, the number of cell colonies in miR-148a-3p overexpression or MAP3K9 knockdown groups was significantly lower than that in the control groups (Fig. 4c).
Subsequently, we evaluated the function of miR-148a-3p and MAP3K9 in the migration and invasion of LUAD cells. In contrast to the control groups, miR-148a-3p mimic or si-MAP3K9 significantly inhibited the cell invasion and migration (Fig. 5a and b).
a and b Transwell assay was applied to detect the invasion and migration of miR-148a-3p overexpressed or MAP3K9 knockdown cells. c F-actin was stained with rhodamine phalloidin, and nuclei were stained with DAPI. d The protein levels of EMT markers in miR-148a-3p overexpressed or MAP3K9 knockdown cells were determined (n = 3). **P < 0.01 and ***P < 0.001.
To further show the reversal of EMT in LUAD cells, we observed the morphology of A549 and NCI-H1299 cells using confocal microscopy. The typical morphology consisting of filopodial structures and prominent lamellipodia was observed in the untreated cells. Importantly, filopodium formation and impaired F-actin polymerization were observed in LUAD cells after transfection with the miR-148a-3p mimic or si-MAP3K9 (Fig. 5c). We further tested the role of MAP3K9 in miR-148a-3p-induced EMT phenotypes. Treatment with the miR-148a-3p mimic or si-MAP3K9 inhibited the expression levels of EMT markers, including N-cadherin, vimentin, and Snail, and consistently increased E-cadherin levels in LUAD (A549 and NCI-H1299) cells (Fig. 5d). Based on the above results, we concluded that miR-148a-3p regulated the cell proliferation and metastasis by targeting MAP3K9.
Tumor growth is inhibited by miR-148a-3p in vivo
To explore the function of miR-148a-3p in tumorigenesis in vivo, the anti-tumor effect of miR-148a-3p was tested using NCI-H1299 and NCI-H1299miR-148a-3p tumor-bearing mice. Entranster reagent, a nano-polymer transfection reagent that can encapsulate nucleic acids into nanoparticles, was employed to encapsulate miR-148a-3p for in vivo delivery with high efficiency and low toxicity. The relative tumor volumes of the encapsulated miR-148a-3p and NCI-H1299miR-148a-3p groups were approximately threefold smaller than that of the saline + NCI-H1299 group (Fig. 6a and b). Consistently, the tumor weights were significantly reduced in the encapsulated miR-148a-3p + NCI-H1299 and NCI-H1299miR-148a-3p groups (Fig. 6c). The free, encapsulated, or overexpression miR-148a-3p stable cells showed no toxic effects, as they had no influence on the body weights of the mice compared to the saline-treated group (Fig. 6d). Moreover, the apoptosis rate of the tumor tissues was evaluated using TUNEL staining. Our results revealed that encapsulated or overexpression miR-148a-3p stable cells had a higher rate of apoptosis than the saline and free miR-148a-3p groups (Fig. 6e). Meanwhile, RT-qPCR results indicated that encapsulated miR-148a-3p significantly inhibited MAP3K9 expression and upregulated miR-148a-3p levels (Fig. 6f and g). Similar results were observed in the stable cells overexpressing miR-148a-3p. Western blotting results also demonstrated that MAP3K9 protein levels were downregulated in the encapsulated miR-148a-3p and NCI-H1299miR-148a-3p groups (Fig. 6h).
The NCI-H1299 cells or NCI-H1299 cells stably overexpressing miR‑148a-3p (NCI-H1299miR-148a-3p) were injected into the right flank regions of mice. Fourteen days later, the mice injected with NCI-H1299 cells were randomly divided into three groups for further treatment as indicated. a The excised tumors were photographed. b The changes of the tumor volumes were determined. c The excised tumors were weighed. d The changes of the body weight. e TUNEL staining of the tumor tissues. The mRNA levels of MAP3K9 (f) and miR-148a-3p (g) were detected in tumors. h The protein levels of MAP3K9 were measured in tumors. *P < 0.05, **P < 0.01 and ***P < 0.001.
The structure of the tumor tissues was analyzed using H&E staining. In the saline-treated group, regular shapes and complete structures without necrotic areas were observed in the solid tumor sections. In contrast, in the encapsulated miR-148a-3p and NCI-H1299miR-148a-3p groups, an increase in the necrotic area in the cell architecture was observed (Supplementary Fig. S4). Histological characterization of the isolated organs was performed to assess their toxicity. No noticeable signs of damage were observed in the lungs, spleen, kidneys, heart, and liver, suggesting that free miR-148a-3p, encapsulated miR-148a-3p, or overexpression miR-148a-3p stable cells showed no significant toxicity in mice (Supplementary Fig. S4). These data indicate that upregulation of miR-148a-3p inhibits lung cancer cell growth in vivo.
Discussion
Abnormal miRNA expression has been observed in various cancers and is known to contribute to tumorigenesis and disease progression. miR-148a-3p has been identified as a tumor suppressor in various types of human malignant cancers, such as NSCLC and colorectal, gastric, ovarian, and breast cancers [8, 19,20,21,22,23,24,25]. In our microarray assay, miR-148a-3p was one of the most downregulated miRNAs among the differentially regulated miRNAs in LUAD tissues. We further confirmed the downregulation of miR-148a-3p in 56 sets of LUAD tumor tissues and in lung cancer cells (A549, NCI-H1299, and NCI-H460). We found that low expression of miR-148a-3p was associated with worse overall survival of patients with LUAD, indicating that it may be a prognostic factor for LUAD.
DNA methylation is one of the most important epigenetic modifications necessary for cellular functions [18]. Aberrant promoter methylation may lead to tumorigenesis [26]. Some miRNAs are tumor suppressors, but are usually silenced due to DNA hypermethylation of their promoters in cancer cells and tissues [27]. In our study, we found that the methylation levels of the miR-148a-3p promoter were notably increased in lung cancer cells and tissues; however, treatment with 5-Aza restored the miR-148a-3p expression. These results suggest that promoter hypermethylation represses miR-148a-3p. Our results confirmed that the methylation and subsequent downregulation of miR-148a-3p have an impact on lung tumorigenesis.
Aberrant expression of DNMTs is closely associated with various types of cancer, as it leads to the silencing of certain tumor suppressor genes [28, 29]. Previous studies reported that DNMT1 contributes to the hypermethylation of the miR-148a-3p promoter. Conversely, miR-148a-3p suppresses DNMT1 expression by binding to the 3′-UTR of DNMT1 mRNA [30, 31]. In the present study, the expression levels of DNMT1 were found to be significantly upregulated in LUAD cells, which could be a possible reason for the epigenetic inactivation of miR-148a-3p. Consistently, inhibition of DNMT1 by 5-Aza or siRNA decreased the CpG island methylation levels in the miR-148a-3p promoter, thereby increasing the miR-148a-3p levels. On the other hand, hypermethylation of miR-148a-3p in LUAD cancer could lead to increased expression of DNMT1, suggesting a mutual negative feedback loop between DNMT1 and miR-148a-3p.
Moreover, we first investigated the correlation between miR-148a-3p expression and its methylation status in clinical lung cancer tissues. Interestingly, we found a significant reverse correlation between methylation and the expression levels of miR-148a-3p, confirming that miR-148a-3p was downregulated in LUAD due to promoter hypermethylation. Metastasis is the foremost cause of cancer morbidity and mortality, and is responsible for ~90% of cancer-related deaths [32]. Tumor nodes and infiltration of malignant cells are the main characteristics of tumor invasion and metastasis. In the metastatic group, the levels of miR-148a-3p were significantly lower than those in the non-metastatic group. Moreover, the results of statistical tests demonstrated that the promoter methylation rates of miR-148a-3p in the lymph node metastasis and no lymph node metastasis subgroups reached a statistically significant difference. These results indicate a significant association between miR-148a-3p methylation and the status of tumor metastasis, including tumor cell infiltration and number of tumor nodes. The promoter methylation-associated downregulation of miR-148a-3p contributed to the metastasis of lung cancer, highlighting that either the expression or methylation status of miR-148a-3p might act as prognostic/predictive markers in LUAD.
MAP3K9 mutations have been observed in various types of human malignant cancers, including metastatic melanoma [33], lung cancer [34], salivary gland malignancy [35], and colon cancer [36]. MAP3K9 is an oncogene regulated by miR-7 that suppresses the progression of pancreatic cancer [37]. It is also the target gene of miR-148a in the development of squamous cell carcinoma [38, 39]. Consistent with previous reports, we confirmed MAP3K9 as a direct target of miR-148a-3p and found a negative correlation between miR-148a-3p and MAP3K9 mRNA levels in patients with LUAD. Our present study revealed that miR-148a-3p targeted MAP3K9 directly, and downregulation of miR-148a-3p resulted in MAP3K9 upregulation. To clarify the role of MAP3K9 in LUAD, its expression levels were measured in clinical tissues and lung cancer cells. We observed higher MAP3K9 levels in LUAD tumor tissues than in normal tissues. Moreover, treatment with 5-Aza or si-DNMT1 inhibited MAP3K9 expression. In vitro results showed that miR-148a-3p overexpression or silencing of MAP3K9 inhibited tumor growth and metastasis by regulating the cell proliferation, colony formation, migration, and invasion. Our findings indicate that DNA methylation-associated downregulation of miR-148a-3p could contribute to the metastasis of LUAD by targeting MAP3K9.
EMT is involved in tumor development and promotes cell proliferation, invasion, and metastasis [40]. EMT is accompanied by changes in epithelial markers, such as E-cadherin, N-cadherin, vimentin, and Snail [41,42,43]. miR-148a-3p overexpression or silencing of MAP3K9 upregulated the E-cadherin protein levels and inhibited N-cadherin, vimentin, and Snail levels. During EMT progression, the migratory and invasive properties are partially induced by reorganization of the actin cytoskeleton [44, 45]. We found that miR-148a-3p overexpression or MAP3K9 knockdown inhibited the formation of invadopodia and reorganization of the cytoskeleton. Therefore, we hypothesized that the miR-148a-3p/MAP3K9 axis regulates EMT, at least partially, by regulating cytoskeleton reorganization.
Consistently, in vivo models indicated that tumor growth was significantly inhibited by the upregulation of miR-148a-3p and downregulation of MAP3K9 in the encapsulated miR-148a-3p and NCI-H1299miR-148a-3p groups. These results suggest that miR-148a-3p contributes to the inhibition of tumor growth via MAP3K9 in LUAD.
In summary, our present study revealed that miR-148a-3p inhibited tumor growth by silencing MAP3K9 expression in lung cancer and its expression was restored by promoter demethylation, indicating that the miR-148a-3p/MAP3K9 axis may act as an effective therapeutic target for lung cancer treatment (Fig. 7).
References
Quintanal-Villalonga A, Molina-Pinelo S. Epigenetics of lung cancer: a translational perspective. Cell Oncol (Dordr). 2019;42:739–56.
van Kampen JGM, van Hooij O, Jansen CF, Smit FP, van Noort PI, Schultz I, et al. miRNA-520f reverses epithelial to mesenchymal transition by targeting ADAM9 and TGFBR2. Cancer Res. 2017;77:2008–17.
Wang FF, Meng F, Wang LL, Wong SC, Cho WC, Chan LW. Associations of mRNA: microRNA for the shared downstream molecules of EGFR and alternative tyrosine tinase receptors in non-small cell lung cancer. Front Genet. 2016;7:173.
Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–76.
Ma N, Zhang WH, Qiao CH, Luo H, Zhang X, Liu DH, et al. The tumor suppressive role of miRNA-509-5p by targeting FOXM1 in non-small cell lung cancer. Cell Physiol Biochem. 2016;38:1435–46.
Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52.
Zhao W, Zheng JB, Wei GB, Yang K, Wang GH, Sun XJ. miR-148a inhibits cell proliferation and migration through targeting ErbB3 in colorectal cancer. Oncol Lett. 2019;18:2530–6.
Li J, Yu T, Cao J, Liu L, Liu Y, Kong HW, et al. MicroRNA-148a suppresses invasion and metastasis of human non-small-cell lung cancer. Cell Physiol Biochem. 2015;37:1847–56.
Slattery ML, Lundgreen A, Wolff RK. Dietary influence on MAPK-signaling pathways and risk of colon and rectal cancer. Nutr Cancer. 2013;65:729–38.
Hannes S, Abhari BA, Fulda S. Smac mimetic triggers necroptosis in pancreatic carcinoma cells when caspase activation is blocked. Cancer Lett. 2016;380:31–8.
Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, Stallings RL. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;11:33.
Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179–83.
Kerr KM, Galler JS, Hagen JA, Laird PW, Laird-Offringa IA. The role of DNA methylation in the development and progression of lung adenocarcinoma. Dis Markers. 2007;23:5–30.
Liang L, Fu JJ, Wang SR, Cen HY, Zhang LM, Mandukhail SR, et al. MiR-142-3p enhances chemosensitivity of breast cancer cells and inhibits autophagy by targeting HMGB1. Acta Pharm Sin B. 2020;10:1036–46.
Xin L, Liu L, Liu C, Zhou LQ, Zhou Q, Yuan YW, et al. DNA-methylation-mediated silencing of miR-7-5p promotes gastric cancer stem cell invasion via increasing Smo and Hes1. J Cell Physiol. 2020;235:2643–54.
Chen WJ, Tu Q, Yu L, Xu YY, Yu G, Jia BL, et al. LncRNA ADAMTS9-AS1, as prognostic marker, promotes cell proliferation and EMT in colorectal cancer. Hum Cell. 2020;33:1133–41.
Xiao J, Liu YK, Wu FX, Liu RY, Xie YL, Yang Q, et al. miR-639 expression is silenced by DNMT3A-mediated hypermethylation and functions as a tumor suppressor in liver cancer cells. Mol Ther. 2020;28:587–98.
Zhang L, Xing ML, Wang XG, Cao WH, Wang HB. MiR-148a-3p suppresses invasion and induces apoptosis of breast cancer cells by regulating USP4 and BIM expression. Int J Clin Exp Pathol. 2017;10:8361–8.
Yu BQ, Lv X, Su LP, Li JF, Yu YY, Gu QL, et al. MiR-148a-3p functions as a tumor suppressor by targeting CCK-BR via inactivating STAT3 and Akt in human gastric cancer. PLoS ONE. 2016;11:e158961.
Zhang J, Ying ZZ, Tang ZL, Long LQ, Li K. MicroRNA-148a-3p promotes myogenic differentiation by targeting the ROCK1 gene. J Biol Chem. 2012;287:21093–101.
Song H, Wang Q, Wen JG, Liu SN, Gao XS, Cheng J, et al. ACVR1, a therapeutic target of fibrodysplasia ossificans progressiva, is negatively regulated by miR-148a-3p. Int J Mol Sci. 2012;13:2063–77.
Zheng BQ, Liang LH, Wang CM, Huang SL, Cao X, Zha RP, et al. MicroRNA-148a-3p suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–83.
Xie Q, Yu ZP, Lu Y, Fan JY, Ni YM, Ma L. microRNA-148a-3p inhibited the proliferation and epithelial–mesenchymal transition progression of non-small-cell lung cancer via modulating Ras/MAPK/Erk signaling. J Cell Physiol. 2019;234:12786–99.
He M, Xue Y. MicroRNA-148a suppresses proliferation and invasion potential of non-small cell lung carcinomas via regulation of STAT3. Onco Targets Ther. 2017;10:1353–61.
Jones P, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92.
Ma L, Teruya-Feldstein J, Weinberg R. Tumor invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8.
Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–22.
Jin F, Dowdy SC, Xiong Y, Eberhardt NL, Podratz KC, Jiang SW. Up-regulation of DNA methyltransferase 3B expression in endometrial cancers. Gynecol Oncol. 2005;96:531–8.
Li X, Wang LH, Cao XC, Zhou LL, Xu C, Cui YH, et al. Casticin inhibits stemness of hepatocellular carcinoma cells via disrupting the reciprocal negative regulation between DNMT1 and miR-148a-3p. Toxicol Appl Pharmacol. 2020;396:114998.
Wang XX, Zhang HY, Li Y. Preliminary study on the role of miR‑148a and DNMT1 in the pathogenesis of acute myeloid leukemia. Mol Med Rep. 2019;19:2943–52.
Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8.
Stark MS, Woods SL, Gartside MG, Bonazzi VF, DuttonRegester K, Aoude LG, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet. 2011;44:165–9.
Fawdar S, Trotter EW, Li YY, Stephenson NL, Hanke F, Marusiak AA, et al. Targeted genetic dependency screen facilitates identification of actionable mutations in FGFR4, MAP3K9, and PAK5 in lung cancer. Proc Natl Acad Sci USA. 2013;110:12426–31.
Kang H, Tan M, Bishop JA, Jones S, Sausen M, Ha PK, et al. Whole-exome sequencing of salivary gland mucoepidermoid carcinoma. Clin Cancer Res. 2017;23:283–8.
Slattery ML, Lundgreen A, Wolff RK. MAP kinase genes and colon and rectal cancer. Carcinogenesis 2012;33:2398–408.
Nie F, Liu TM, Zhong L, Yang XG, Liu YH, Xia HW, et al. MicroRNA-148b enhances proliferation and apoptosis in human renal cancer cells via directly targeting MAP3K9. Mol Med Rep. 2016;13:83–90.
Luo Q, Li W, Zhao T, Tian X, Liu YM, Zhang XB. Role of miR-148a in cutaneous squamous cell carcinoma by repression of MAPK pathway. Arch Biochem Biophys. 2015;583:47–54.
Zhang BX, Yu T, Yu Z, Yang XG. MicroRNA-148a regulates the MAPK/ERK signaling pathway and suppresses the development of esophagus squamous cell carcinoma via targeting MAP3K9. Eur Rev Med Pharmacol Sci. 2019;23:6497–504.
Gao W, Zhang CM, Li WQ, Li HZ, Sang JW, Zhao QL, et al. Promoter methylation-regulated miR-145-5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol Ther. 2019;27:365–79.
Landeros NL, Santoro PM, Carrasco-Avino G, Corvalan AH. Competing endogenous RNA networks in the epithelial to mesenchymal transition in diffuse-type of gastric cancer. Cancers (Basel). 2020;12:2741.
Zhang X, Bai J, Yin H, Long L, Zheng ZW, Wang QQ, et al. Exosomal miR-1255b-5p targets human telomerase reverse transcriptase in colorectal cancer cells to suppress epithelial-to-mesenchymal transition. Mol Oncol. 2020;14:2589–608.
Camerlingo R, Miceli R, Marra L, Rea G, Agnano I, D’Nardella M, et al. Conditioned medium of primary lung cancer cells induces EMT in A549 lung cancer cell line by TGF-β1 and miRNA21 cooperation. PLoS ONE. 2019;14:e219597.
Izdebska M, Zielinska W, Grzanka D, Gagat M. The role of actin dynamics and actin-binding proteins expression in epithelial-to-mesenchymal transition and its association with cancer progression and evaluation of possible therapeutic targets. BioMed Res Int. 2018;2018:4578373.
Peng JM, Bera R, Chiou CY, Yu MC, Chen TC, Chen CW, et al. Actin cytoskeleton remodeling drives epithelial-mesenchymal transition for hepatoma invasion and metastasis in mice. Hepatology. 2018;67:2226–43.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 82000080, 82070406, 81330007 and U1601227), Guangzhou Science and Technology Plan Project (202102020073) and traditional Chinese medicine Program of Guangdong (20211240). This work was also supported by the Department of Science and Technology of Guangdong (2020A1515011158), the Department of Education of Guangdong (2020KTSCX106) and the High-level University Construction Fund of Guangdong (06-410-2107210).
Author information
Authors and Affiliations
Contributions
LL, WYX, HYC, YYQ, LT, LMZ, YZ, and ZJC conceived, designed, and interpreted the study. JJF, APQ, XPL, and SPL undertook the data acquisition and analysis. XYY, AS, and JHH were responsible for the comprehensive technical support. LL, AS, and WYX were major contributors in writing the paper. LL and WYX contributed to the inspection of data and final paper. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Liang, L., Xu, Wy., Shen, A. et al. Promoter methylation-regulated miR-148a-3p inhibits lung adenocarcinoma (LUAD) progression by targeting MAP3K9. Acta Pharmacol Sin 43, 2946–2955 (2022). https://doi.org/10.1038/s41401-022-00893-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-022-00893-8