Featured
-
-
Letter |
 Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit
Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit
A sub-nanometre reconstruction of a 40S complex containing eIF3 and a hepatitis C virus (HCV)-like internal ribosome entry site (IRES) shows that the IRES displaces eIF3 from the 40S and sequesters it to gain access to the 40S subunit.
- Yaser Hashem
- , Amedee des Georges
- & Joachim Frank
-
Article |
 The initiation of mammalian protein synthesis and mRNA scanning mechanism
The initiation of mammalian protein synthesis and mRNA scanning mechanism
Three structures of the eukaryotic small ribosomal subunit in complex with initiator tRNA, mRNA and the initiation factors eIF1 and eIF1A have been solved; these structures offer insight into the contributions of the initiation factors, the mechanism by which mRNA is scanned, and the interactions that occur in the ribosome’s P site.
- Ivan B. Lomakin
- & Thomas A. Steitz
-
Letter |
 Unusual base pairing during the decoding of a stop codon by the ribosome
Unusual base pairing during the decoding of a stop codon by the ribosome
Here, the structure of the 30S ribosomal subunit and the 70S ribosome in complex with a messenger RNA with pseudouridine in the place of uridine reveals unexpected base pairing.
- Israel S. Fernández
- , Chyan Leong Ng
- & V. Ramakrishnan
-
Letter |
 Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Members of the SAM-dependent methyltransferase superfamily are involved in the modification of wobble uridine to 5-oxacetyl uridine in Gram-negative bacteria; CmoA converts SAM to carboxy-SAM (Cx-SAM; a metabolite that was unknown previously), and CmoB uses Cx-SAM to convert 5-hydroxyuridine to 5-oxyacetyl uridine in tRNA.
- Jungwook Kim
- , Hui Xiao
- & Steven C. Almo
-
Article |
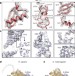 Structures of the human and Drosophila 80S ribosome
Structures of the human and Drosophila 80S ribosome
High-resolution cryo-EM density maps are used to present the structures of Drosophila and human 80S ribosomes in complex with eEF2, E-site transfer RNA and Stm1-like proteins, and reveal the presence of two additional structural layers in the ribosomes of metazoan eukaryotes.
- Andreas M. Anger
- , Jean-Paul Armache
- & Roland Beckmann
-
Letter |
 Non-optimal codon usage affects expression, structure and function of clock protein FRQ
Non-optimal codon usage affects expression, structure and function of clock protein FRQ
The frq gene, essential for circadian clock function, is shown to differ from most other genes in Neurospora by exhibiting non-optimal codon usage; by contrast, optimization of codon usage is unexpectedly found to affect the structure and function of the coded protein, subsequently impairing circadian feedback loops.
- Mian Zhou
- , Jinhu Guo
- & Yi Liu
-
Letter |
 Non-optimal codon usage is a mechanism to achieve circadian clock conditionality
Non-optimal codon usage is a mechanism to achieve circadian clock conditionality
Central circadian proteins in cyanobacteria unexpectedly use non-optimal codons, and optimizing their codes is shown to cause a change in an adaptive response to environmental conditions.
- Yao Xu
- , Peijun Ma
- & Carl Hirschie Johnson
-
Letter |
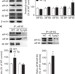 Exaggerated translation causes synaptic and behavioural aberrations associated with autism
Exaggerated translation causes synaptic and behavioural aberrations associated with autism
Mice overexpressing eIF4E show autism-related behaviours and altered synaptic activity in the hippocampus, prefrontal cortex and striatum, and these phenotypes can be rescued with the cap-dependent translation inhibitor 4EGI-1.
- Emanuela Santini
- , Thu N. Huynh
- & Eric Klann
-
Research Highlights |
RNA tails time protein production
-
News & Views |
 All clear for ribosome landing
All clear for ribosome landing
The discovery of a dramatic structural rearrangement that is stabilized by an RNA scaffold helps to explain how nascent proteins are delivered for export from the cell cytoplasm. See Letter p.271
- Harris D. Bernstein
-
News |
 Autism symptoms reversed in mice
Autism symptoms reversed in mice
Neural 'hyperconnections' caused by runaway protein production can be undone.
- Dan Jones
-
Article |
 Autism-related deficits via dysregulated eIF4E-dependent translational control
Autism-related deficits via dysregulated eIF4E-dependent translational control
Mice lacking 4E-BP2, an eIF4E repressor, display increased translation of neuroligins; the mice also show autism-related behaviours and alterations in hippocampal synaptic activity, and these are reversed by normalization of eIF4E activity or neuroligin 1 levels.
- Christos G. Gkogkas
- , Arkady Khoutorsky
- & Nahum Sonenberg
-
Letter |
 Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat
Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat
Antisense Uchl1, a long non-coding RNA that is an antisense transcript for the Uchl1 gene, upregulates UCHL1 protein levels through the combined action of an overlapping sequence at its 5′ end and an embedded SINEB2 element.
- Claudia Carrieri
- , Laura Cimatti
- & Stefano Gustincich
-
Letter |
 Heterogeneous pathways and timing of factor departure during translation initiation
Heterogeneous pathways and timing of factor departure during translation initiation
A single-molecule approach is used to investigate the kinetics of assembly of the translation initiation complex, revealing that there is more than one pathway by which the necessary factors can assemble.
- Albert Tsai
- , Alexey Petrov
- & Joseph D. Puglisi
-
Letter |
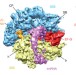 The complex of tmRNA–SmpB and EF-G on translocating ribosomes
The complex of tmRNA–SmpB and EF-G on translocating ribosomes
Stalled bacterial ribosomes can be rescued by interaction with SmpB protein and a highly structured transfer-messenger RNA, and a cryo-electron microscopy map of this complex now shows how EF-G-dependent translocation of this non-canonical ligand is facilitated by conformational changes in the ribosome and the transfer-messenger RNA.
- David J. F. Ramrath
- , Hiroshi Yamamoto
- & Christian M. T. Spahn
-
Letter |
 An oxygen-regulated switch in the protein synthesis machinery
An oxygen-regulated switch in the protein synthesis machinery
Hypoxia activates a translation initiation pathway that escapes global inhibition of protein synthesis.
- James Uniacke
- , Chet E. Holterman
- & Stephen Lee
-
Letter |
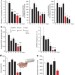 Sustained translational repression by eIF2α-P mediates prion neurodegeneration
Sustained translational repression by eIF2α-P mediates prion neurodegeneration
Accumulation of prion protein during prion replication causes persistent translational repression of global protein synthesis, which is mediated by eIF2α-P and is associated with synaptic failure and neuronal loss in prion-diseased mice; promoting translational recovery in hippocampi of prion-infected mice is neuroprotective.
- Julie A. Moreno
- , Helois Radford
- & Giovanna R. Mallucci
-
News & Views |
 The director's cut
The director's cut
A genome-wide characterization of active translation of messenger RNA following inhibition of mTOR will transform our view of this signalling protein's regulatory role in cancer. See Article p.55 & Letter p.109
- Antonio Gentilella
- & George Thomas
-
Letter |
 A unifying model for mTORC1-mediated regulation of mRNA translation
A unifying model for mTORC1-mediated regulation of mRNA translation
mTORC1 is shown to regulate a translational program that requires the rapamycin-resistant 4E-BP family of translational repressors and consists almost entirely of mRNAs containing 5′ terminal oligopyrimidine or related motifs.
- Carson C. Thoreen
- , Lynne Chantranupong
- & David M. Sabatini
-
Letter |
 The anti-Shine–Dalgarno sequence drives translational pausing and codon choice in bacteria
The anti-Shine–Dalgarno sequence drives translational pausing and codon choice in bacteria
Internal Shine–Dalgarno-like sequences in bacterial messenger RNA determine the elongation rate of protein synthesis and synonymous codon usage.
- Gene-Wei Li
- , Eugene Oh
- & Jonathan S. Weissman
-
Letter |
 A new understanding of the decoding principle on the ribosome
A new understanding of the decoding principle on the ribosome
An integrated mechanism for decoding is proposed, based on six X-ray structures of the 70S ribosome determined at 3.1–3.4 Å resolution, modelling cognate or near-cognate states of the decoding centre at the proofreading step.
- Natalia Demeshkina
- , Lasse Jenner
- & Gulnara Yusupova
-
Article |
 The translational landscape of mTOR signalling steers cancer initiation and metastasis
The translational landscape of mTOR signalling steers cancer initiation and metastasis
Ribosome profiling reveals specialized translation of the prostate cancer genome by oncogenic mTOR signalling; stringent inhibition of mTOR signalling reduces prostate cancer invasion and metastasis in a mouse model.
- Andrew C. Hsieh
- , Yi Liu
- & Davide Ruggero
-
Article |
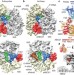 Structural basis of highly conserved ribosome recycling in eukaryotes and archaea
Structural basis of highly conserved ribosome recycling in eukaryotes and archaea
Cryo-electron-microscopy reconstructions of eukaryotic and archaeal ribosomes bound by ABCE1 and Pelota suggest a conserved mechanism for ribosome recycling.
- Thomas Becker
- , Sibylle Franckenberg
- & Roland Beckmann
-
Letter |
 An equilibrium-dependent retroviral mRNA switch regulates translational recoding
An equilibrium-dependent retroviral mRNA switch regulates translational recoding
The NMR structure of the murine leukaemia virus recoding translational signal is determined, giving insight into the mechanism of read-through in retroviruses.
- Brian Houck-Loomis
- , Michael A. Durney
- & Victoria M. D’Souza
-
Letter |
 Different substrate-dependent transition states in the active site of the ribosome
Different substrate-dependent transition states in the active site of the ribosome
- Stephan Kuhlenkoetter
- , Wolfgang Wintermeyer
- & Marina V. Rodnina
-
Letter |
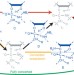 A two-step chemical mechanism for ribosome-catalysed peptide bond formation
A two-step chemical mechanism for ribosome-catalysed peptide bond formation
- David A. Hiller
- , Vipender Singh
- & Scott A. Strobel
-
Letter |
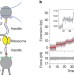 The ribosome uses two active mechanisms to unwind messenger RNA during translation
The ribosome uses two active mechanisms to unwind messenger RNA during translation
- Xiaohui Qu
- , Jin-Der Wen
- & Ignacio Tinoco
-
News & Views |
 Stop the nonsense
Stop the nonsense
A subtle biochemical alteration can reprogram signals that herald the termination of protein translation into signals encoding amino acids at the level of messenger RNA — and without altering the corresponding DNA. See Letter p.395
- Adrian R. Ferré-D'Amaré
-
Letter |
 Converting nonsense codons into sense codons by targeted pseudouridylation
Converting nonsense codons into sense codons by targeted pseudouridylation
- John Karijolich
- & Yi-Tao Yu
-
Technology Feature |
 Real-time analysis
Real-time analysis
-
Letter |
 Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites
Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites
During translation, tRNAs enter the ribosome and then move sequentially through three sites, known as A, P and E, as they transfer their attached amino acids onto the growing peptide chain. How the ribosome facilitates tRNA translocation between the sites remains largely unknown. Now a study uses multiparticle cryoelectron microscopy of a ribosome bound to the translation elongation factor, EF-G, to get information about tRNA movement. It identifies two new substates and sees that translocation is linked to unratcheting of the 30S ribosomal subunit.
- Andreas H. Ratje
- , Justus Loerke
- & Christian M. T. Spahn
-
News |
 Elusive protein factory mapped at last
Elusive protein factory mapped at last
Victory claimed in race to determine structure of eukaryotic ribosome.
- Daniel Cressey
-
Article |
 Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA
Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA
tRNAs are synthesized in a premature form that requires trimming of the 5′ and 3′ ends and modification of specific nucleotides. RNase P, a complex containing a long catalytic RNA and a protein cofactor, catalyses the cleavage that generates the mature 5′ end. Here, the structure of RNase P bound to mature tRNAPhe is solved. Recognition of the leader sequence and its mechanism of cleavage is determined by soaking an oligonucleotide corresponding to the premature 5′ end into the crystal.
- Nicholas J. Reiter
- , Amy Osterman
- & Alfonso Mondragón
-
News |
 DNA sequence may be lost in translation
DNA sequence may be lost in translation
Researchers tackle a mysterious genetic phenomenon
- Alla Katsnelson
-
Letter |
 Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions
Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions
In most bacteria and all archaea, glutamyl-tRNA synthetase (GluRS) glutamylates both tRNAGlu and tRNAGln; Glu-tRNAGln is then converted to Gln-tRNAGln by an amidotransferase. Here the structure is reported of a bacterial complex containing tRNAGln, GluRS and the amidotransferase GatCAB. The structure provides an explanation for how the enzymes work consecutively: only one can assume a productive state at any time. There also seems to be an intermediary state in which neither enzyme is productive.
- Takuhiro Ito
- & Shigeyuki Yokoyama
-
Research Highlights |
Molecular biology: Proteins actin' differently
-
Letter |
 Role of a ribosome-associated E3 ubiquitin ligase in protein quality control
Role of a ribosome-associated E3 ubiquitin ligase in protein quality control
The translation of messenger RNA that lacks stop codons results in the production of aberrant proteins, which may have harmful effects on the cell. It is unclear how eukaryotic cells eliminate these 'non-stop' proteins. Here it is shown that, in Saccharomyces cerevisiae, an E3 ubiquitin ligase called Ltn1 acts in the quality-control pathway. It associates with ribosomes and marks non-stop proteins with ubiquitin, which targets the proteins for degradation.
- Mario H. Bengtson
- & Claudio A. P. Joazeiro
-
Letter |
 A ribosome-associating factor chaperones tail-anchored membrane proteins
A ribosome-associating factor chaperones tail-anchored membrane proteins
Tail-anchored proteins have a single transmembrane domain at their carboxy termini and are post-translationally targeted to the endoplasmic reticulum via the cytosolic ATPase TRC40. These authors identify a conserved protein complex called Bat3 complex that is recruited to ribosomes, interacts with the transmembrane domain of newly released tail-anchored proteins and transfers them to TRC40 for subsequent targeting to the endoplasmic reticulum.
- Malaiyalam Mariappan
- , Xingzhe Li
- & Ramanujan S. Hegde
-
Letter |
 New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism
New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism
In the course of characterizing short RNAs from human cells using single-molecule high-throughput sequencing, these authors identify a new short RNA species. The presence of non-genomically encoded poly(U) residues at their 5' ends implies the existence of an unknown RNA copying mechanism in human cells.
- Philipp Kapranov
- , Fatih Ozsolak
- & Patrice M. Milos
-
Letter |
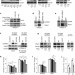 Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression
Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression
LRRK2 activity is dysregulated in Parkinson's disease, but how it contributes to the pathogenesis is unknown. These authors show that Drosophila LRRK2 interacts with the Argonaute component (dAgo1) of the RNA-induced silencing complex. This is associated with reduced levels of dAgo1, interaction between phospho-4E-BP1 and hAgo2, and impairment of microRNA-mediated repression. This leads to overexpression of the cell cycle genes e2f1 and dp, and consequent degeneration of dopaminergic neurons.
- Stephan Gehrke
- , Yuzuru Imai
- & Bingwei Lu
-
Article |
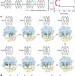 Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy
Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy
During protein synthesis within the ribosome, transfer RNAs (tRNAs) move sequentially through different sites as their attached amino acids are transferred onto the growing protein chain. Large conformational movements accompany this process. Here, a staggering 1.9 million electron cryomicroscopy images of the ribosome have been processed to visualize these changes. The results reveal that the ribosome functions as a Brownian machine that couples spontaneous changes driven by thermal energy to directed movement.
- Niels Fischer
- , Andrey L. Konevega
- & Holger Stark
-
News & Views |
 Translocation in slow motion
Translocation in slow motion
Time-resolved electron microscopy can capture structural changes in active macromolecular complexes, but detailed imaging is essential. The dynamics of one step in protein synthesis has been deduced from two million images.
- Måns Ehrenberg
-
Letter |
 Principles of stop-codon reading on the ribosome
Principles of stop-codon reading on the ribosome
Stop codons in messenger RNA define when a protein sequence has been completely synthesized; such codons bind release factors (RFs), which cause the newly made protein to be released. Structures of RFs alone and in combination with the ribosome have been reported, but the energetics of the reaction in the presence of codons had not been determined. Here, molecular dynamics simulations of 14 termination complexes are used to define how termination is achieved and how the RFs distinguish different sequences.
- Johan Sund
- , Martin Andér
- & Johan Åqvist
-
Letter |
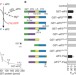 eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation
eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation
The initiation of protein synthesis requires the eukaryotic translation initiation factor (eIF) 2, which uses energy from the hydrolysis of GTP. Another factor, eIF5, accelerates the GTP-hydrolysing activity of eIF2. Here, two other roles for eIF5 have been defined. One involves stabilizing GDP, the product of GTP hydrolysis, on eIF2. In its other role, eIF5 works with phosphorylated eIF2 to inhibit the guanine-nucleotide exchange factor eIF2B. These results clarify our understanding of how the initiation of translation is regulated.
- Martin D. Jennings
- & Graham D. Pavitt
-
Article |
 Real-time tRNA transit on single translating ribosomes at codon resolution
Real-time tRNA transit on single translating ribosomes at codon resolution
Single-molecule studies allow biological processes to be examined one molecule at a time, as they occur. Here, zero-mode waveguides have been used to concentrate reactions in zeptolitre-sized volumes, making it possible to study real-time translocation by the ribosome. The binding of transfer RNAs (tRNAs) to the ribosome could be followed; the results show that tRNA release from the exit site is uncoupled from tRNA binding to the aminoacyl-tRNA site.
- Sotaro Uemura
- , Colin Echeverría Aitken
- & Joseph D. Puglisi
-
News & Views |
 A ribosome in action
A ribosome in action
The manufacture of proteins by ribosomes involves complex interactions of diverse nucleic-acid and protein ligands. Single-molecule studies allow us, for the first time, to follow the synthesis of full-length proteins in real time.
- Susanne Brakmann
-
Letter |
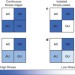 Compensatory evolution in mitochondrial tRNAs navigates valleys of low fitness
Compensatory evolution in mitochondrial tRNAs navigates valleys of low fitness
Evolution from one fitness peak to another must involve either transitions through intermediates of low fitness or skirting round the fitness valley through compensatory mutations elsewhere. Here, the base pairs in mitochondrial tRNA stems is used as a model to show that deep fitness valleys can be traversed. Transitions between AU and GC pairs have occurred during mammalian evolution without help from genetic drift or mutations elsewhere.
- Margarita V. Meer
- , Alexey S. Kondrashov
- & Fyodor A. Kondrashov
-
Letter |
 Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome
Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome
Although new amino acids with desirable properties can be devised, only a few have been successfully introduced into proteins by the cellular machinery. Even then, only one type of unnatural amino acid can be added to a given protein. Here, a new system has been designed that could allow the incorporation of up to 200 novel amino acids. The system involves an orthogonal ribosome that uses quadruplet — rather than triplet — codons, as well as orthogonal tRNA synthetase–tRNA pairs.
- Heinz Neumann
- , Kaihang Wang
- & Jason W. Chin

 Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export
Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export