Abstract
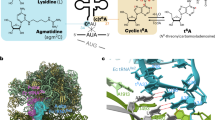
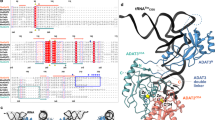
In most bacteria and all archaea, glutamyl-tRNA synthetase (GluRS) glutamylates both tRNAGlu and tRNAGln, and then Glu-tRNAGln is selectively converted to Gln-tRNAGln by a tRNA-dependent amidotransferase1,2. The mechanisms by which the two enzymes recognize their substrate tRNA(s), and how they cooperate with each other in Gln-tRNAGln synthesis, remain to be determined. Here we report the formation of the ‘glutamine transamidosome’ from the bacterium Thermotoga maritima, consisting of tRNAGln, GluRS and the heterotrimeric amidotransferase GatCAB, and its crystal structure at 3.35 Å resolution. The anticodon-binding body of GluRS recognizes the common features of tRNAGln and tRNAGlu, whereas the tail body of GatCAB recognizes the outer corner of the L-shaped tRNAGln in a tRNAGln-specific manner. GluRS is in the productive form, as its catalytic body binds to the amino-acid-acceptor arm of tRNAGln. In contrast, GatCAB is in the non-productive form: the catalytic body of GatCAB contacts that of GluRS and is located near the acceptor stem of tRNAGln, in an appropriate site to wait for the completion of Glu-tRNAGln formation by GluRS. We identified the hinges between the catalytic and anticodon-binding bodies of GluRS and between the catalytic and tail bodies of GatCAB, which allow both GluRS and GatCAB to adopt the productive and non-productive forms. The catalytic bodies of the two enzymes compete for the acceptor arm of tRNAGln and therefore cannot assume their productive forms simultaneously. The transition from the present glutamylation state, with the productive GluRS and the non-productive GatCAB, to the putative amidation state, with the non-productive GluRS and the productive GatCAB, requires an intermediate state with the two enzymes in their non-productive forms, for steric reasons. The proposed mechanism explains how the transamidosome efficiently performs the two consecutive steps of Gln-tRNAGln formation, with a low risk of releasing the unstable intermediate Glu-tRNAGln.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wilcox, M. & Nirenberg, M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl Acad. Sci. USA 61, 229–236 (1968)
Curnow, A. W. et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA 94, 11819–11826 (1997)
Tumbula, D. L., Becker, H. D., Chang, W. Z. & Söll, D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature 407, 106–110 (2000)
Nakamura, A., Yao, M., Chimnaronk, S., Sakai, N. & Tanaka, I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science 312, 1954–1958 (2006)
Oshikane, H. et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science 312, 1950–1954 (2006)
Curnow, A. W., Tumbula, D. L., Pelaschier, J. T., Min, B. & Söll, D. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl Acad. Sci. USA 95, 12838–12843 (1998)
Becker, H. D. & Kern, D. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl Acad. Sci. USA 95, 12832–12837 (1998)
Bailly, M. et al. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 34, 6083–6094 (2006)
Bailly, M., Blaise, M., Lorber, B., Becker, H. D. & Kern, D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell 28, 228–239 (2007)
Ruff, M. et al. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp . Science 252, 1682–1689 (1991)
Sekine, S., Nureki, O., Shimada, A., Vassylyev, D. G. & Yokoyama, S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nature Struct. Biol. 8, 203–206 (2001)
Rampias, T., Sheppard, K. & Söll, D. The archaeal transamidosome for RNA-dependent glutamine biosynthesis. Nucleic Acids Res. (in the press)
Wu, J. et al. Insights into tRNA-dependent amidotransferase evolution and catalysis from the structure of the Aquifex aeolicus enzyme. J. Mol. Biol. 391, 703–716 (2009)
Nakamura, A. et al. Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition. Nucleic Acids Res. 38, 672–682 (2009)
Sekine, S. et al. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. EMBO J. 22, 676–688 (2003)
Ito, T., Kiyasu, N., Matsunaga, R., Takahashi, S. & Yokoyama, S. Crystal structure of nondiscriminating glutamyl-tRNA synthetase from Thermotoga maritima . Acta Crystallogr. D 66, 813–820 (2010)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative. Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
DeLano, W. L. The PyMOL molecular graphics system. (http://www.pymol.org) (2002)
Acknowledgements
We thank S. Sekine for discussions and help with data collection, S. Goto-Ito for reading the manuscript, and S. Takahashi, N. Kiyasu and R. Matsunaga for help with sample preparation. We are grateful to the staff of the beam line BL41XU in the SPring-8 and the beam line AR-NE3A in the Photon Factory for their support during data collection. This work was supported by the Targeted Proteins Research Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and MEXT, and the Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from MEXT.
Author information
Authors and Affiliations
Contributions
T.I. and S.Y. designed the study, T.I. performed the research, and T.I. and S.Y. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Discussion, Supplementary Figures 1-9 with legends, Supplementary Table 1, legend for Supplementary Movie 1 and an additional reference. (PDF 3620 kb)
Supplementary Movie 1
The movie shows the transition of the glutamine transamidsome from the glutamylation state to the amidation state – see Supplementary Movie file for full legend. (MOV 9933 kb)
Rights and permissions
About this article
Cite this article
Ito, T., Yokoyama, S. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature 467, 612–616 (2010). https://doi.org/10.1038/nature09411
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09411
This article is cited by
-
Pathogenic variants in glutamyl-tRNAGln amidotransferase subunits cause a lethal mitochondrial cardiomyopathy disorder
Nature Communications (2018)
-
Structural basis for tRNA-dependent cysteine biosynthesis
Nature Communications (2017)
-
Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes
BMC Genomics (2014)
-
Transfer RNA: A dancer between charging and mis-charging for protein biosynthesis
Science China Life Sciences (2013)
-
TP Atlas: integration and dissemination of advances in Targeted Proteins Research Program (TPRP)—structural biology project phase II in Japan
Journal of Structural and Functional Genomics (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.