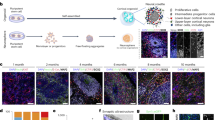
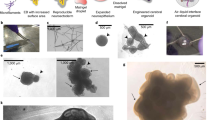
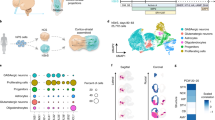
Abstract
The development of neural circuits involves wiring of neurons locally following their generation and migration, as well as establishing long-distance connections between brain regions. Studying these developmental processes in the human nervous system remains difficult because of limited access to tissue that can be maintained as functional over time in vitro. We have previously developed a method to convert human pluripotent stem cells into brain region–specific organoids that can be fused and integrated to form assembloids and study neuronal migration. In contrast to approaches that mix cell lineages in 2D cultures or engineer microchips, assembloids leverage self-organization to enable complex cell–cell interactions, circuit formation and maturation in long-term cultures. In this protocol, we describe approaches to model long-range neuronal connectivity in human brain assembloids. We present how to generate 3D spheroids resembling specific domains of the nervous system and then how to integrate them physically to allow axonal projections and synaptic assembly. In addition, we describe a series of assays including viral labeling and retrograde tracing, 3D live imaging of axon projection and optogenetics combined with calcium imaging and electrophysiological recordings to probe and manipulate the circuits in assembloids. The assays take 3–4 months to complete and require expertise in stem cell culture, imaging and electrophysiology. We anticipate that these approaches will be useful in deciphering human-specific aspects of neural circuit assembly and in modeling neurodevelopmental disorders with patient-derived cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data presented in this protocol are available on request from the corresponding author. Source data are provided with this paper.
References
Winnubst, J. et al. Reconstruction of 1,000 projection neurons reveals new cell types and organization of long-range connectivity in the mouse brain. Cell 179, 268–281.e13 (2019).
Kostovic, I., Rados, M., Kostovic-Srzentic, M. & Krsnik, Z. Fundamentals of the development of connectivity in the human fetal brain in late gestation: from 24 weeks gestational age to term. J. Neuropathol. Exp. Neurol. 80, 393–414 (2021).
Kaiser, T. & Feng, G. Modeling psychiatric disorders for developing effective treatments. Nat. Med. 21, 979–988 (2015).
Pasca, S. P. The rise of three-dimensional human brain cultures. Nature 553, 437–445 (2018).
Velasco, S., Paulsen, B. & Arlotta, P. 3D brain organoids: studying brain development and disease outside the embryo. Annu. Rev. Neurosci. 43, 375–389 (2020).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Bagley, J. A., Reumann, D., Bian, S., Levi-Strauss, J. & Knoblich, J. A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 14, 743–751 (2017).
Xiang, Y. et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21, 383–398.e7 (2017).
Sloan, S. A., Andersen, J., Pasca, A. M., Birey, F. & Pasca, S. P. Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 13, 2062–2085 (2018).
Yoon, S.-J. et al. Reliability of human cortical organoid generation. Nat. Methods 16, 75–78 (2019).
Miura, Y. et al. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 38, 1421–1430 (2020).
Shepherd, G. M. Corticostriatal connectivity and its role in disease. Nat. Rev. Neurosci. 14, 278–291 (2013).
Welch, J. M. et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature 448, 894–900 (2007).
Peca, J. et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472, 437–442 (2011).
Milad, M. R. & Rauch, S. L. Obsessive-compulsive disorder: beyond segregated cortico-striatal pathways. Trends Cogn. Sci. 16, 43–51 (2012).
Sharma, A., Sances, S., Workman, M. J. & Svendsen, C. N. Multi-lineage human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell 26, 309–329 (2020).
Xiang, Y. et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell 24, 487–497.e7 (2019).
Andersen, J. et al. Generation of functional human 3D cortico-motor assembloids. Cell 183, 1913–1929.e26 (2020).
Cakir, B. et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 16, 1169–1175 (2019).
Marton, R. M. et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 22, 484–491 (2019).
Strano, A., Tuck, E., Stubbs, V. E. & Livesey, F. J. Variable outcomes in neural differentiation of human PSCs arise from intrinsic differences in developmental signaling pathways. Cell Rep. 31, 107732 (2020).
Micali, N. et al. Variation of human neural stem cells generating organizer states in vitro before committing to cortical excitatory or inhibitory neuronal fates. Cell Rep. 31, 107599 (2020).
Button, K. S. et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 14, 365–376 (2013).
Pasca, S. P. et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 17, 1657–1662 (2011).
Tian, Y. et al. Alteration in basal and depolarization induced transcriptional network in iPSC derived neurons from Timothy syndrome. Genome Med. 6, 75 (2014).
Sun, Y. et al. A deleterious Nav1.1 mutation selectively impairs telencephalic inhibitory neurons derived from Dravet Syndrome patients. eLife 5, e13073 (2016).
Susaki, E. A. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014).
Tainaka, K. et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 24, 2196–2210.e9 (2018).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
de Leeuw, C. N. et al. rAAV-compatible MiniPromoters for restricted expression in the brain and eye. Mol. Brain 9, 52 (2016).
Chetty, S. et al. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods 10, 553–556 (2013).
Ishikawa, D. et al. Fluorescent pipettes for optically targeted patch-clamp recordings. Neural Netw. 23, 669–672 (2010).
Acknowledgements
We thank members of the Pașca laboratory at Stanford University for experimental support, as well as the Stanford WuTsai Neurosciences Institute Virus Core for production of AAVs. This work was supported by the US National Institutes of Health (NIH) BRAINS Award (MH107800) (to S.P.P.), Stanford Bio-X, the NYSCF Robertson Stem Cell Investigator Award (to S.P.P.), the Stanford Wu Tsai Neurosciences Big Idea Project on Human Brain Organogenesis (to S.P.P.), the Kwan Research Fund (to S.P.P.), the Coates Foundation (to S.P.P.), the Senkut Research Fund (to S.P.P.), the Chan Zuckerberg Initiative Ben Barres Investigator Award (to S.P.P.), the Stanford Medicine Dean’s Postdoctoral Fellowship (to Y.M.) and the Stanford Maternal & Child Health Research Institute (MCHRI) Postdoctoral Fellowship (to Y.M. and O.R.).
Author information
Authors and Affiliations
Contributions
Y.M. collected data and optimized the protocols for generating hStrSs and assembloids, clearing of assembloids, retrograde viral tracing and optogenetics coupled with calcium imaging. M.-Y.L collected data and optimized the protocols for whole-cell recordings. O.R. optimized the protocols and analyzed optogenetics experiments combined with calcium imaging. S.-J.Y. optimized the protocols for generation of hCSs. G.N. collected data and optimized the protocols for clearing of assembloids. Y.M. and S.P.P. wrote the manuscript with input from all authors. S.P.P. supervised this work.
Corresponding author
Ethics declarations
Competing interests
Stanford University has filed and holds patents for the generation of brain region–specific spheroids/organoids and assembloids. Y.M. and S.P.P. are listed as inventors on some of these patents.
Additional information
Peer review information Nature Protocols thanks J. Gray Camp, Madeline A Lancaster and Alysson R. Muotri for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Miura, Y. et al. Nat. Biotechnol. 38, 1421–1430 (2020): https://doi.org/10.1038/s41587-020-00763-w
Yoon, S.-J. et al. Nat. Methods 16, 75–78 (2019): https://doi.org/10.1038/s41592-018-0255-0
Sloan, S. A. et al. Nat. Protoc. 13, 2062–2085 (2018): https://doi.org/10.1038/s41596-018-0032-7
Source data
Source Data Fig. 4
Statistical source data
Rights and permissions
About this article
Cite this article
Miura, Y., Li, MY., Revah, O. et al. Engineering brain assembloids to interrogate human neural circuits. Nat Protoc 17, 15–35 (2022). https://doi.org/10.1038/s41596-021-00632-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00632-z
This article is cited by
-
Laminin-associated integrins mediate Diffuse Intrinsic Pontine Glioma infiltration and therapy response within a neural assembloid model
Acta Neuropathologica Communications (2024)
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
Global Versus Local Theories of Consciousness and the Consciousness Assessment Issue in Brain Organoids
Neuroethics (2024)
-
Humanized brain organoids-on-chip integrated with sensors for screening neuronal activity and neurotoxicity
Microchimica Acta (2024)
-
Microglial over-pruning of synapses during development in autism-associated SCN2A-deficient mice and human cerebral organoids
Molecular Psychiatry (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.