« Prev Next »
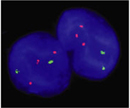
Aneuploidies disturb the delicate balance of gene products in cells. By definition, aneuploid cells have an abnormal number of chromosomes. Because each chromosome contains hundreds of genes, the addition or loss of even a single chromosome disrupts the existing equilibrium in cells, and in most cases, is not compatible with life.
Using the tools of modern cytogenetics, scientists have recently provided new insights into the origins of aneuploidy. Researchers now appreciate that aneuploid gametes are produced at surprisingly high rates in human meioses, and that very few aneuploid embryos are able to survive. Much attention is currently focused on determining how specific imbalances in gene expression lead to the profound phenotypes associated with aneuploid conditions, such as Down syndrome, with the ultimate goal of developing therapeutic interventions.
Most Aneuploidies Are Lethal
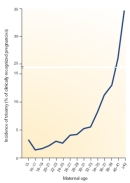
In humans, the most common aneuploidies are trisomies, which represent about 0.3% of all live births. Trisomies are characterized by the presence of one additional chromosome, bringing the total chromosome number to 47. With few exceptions, trisomies do not appear to be compatible with life. In fact, trisomies represent about 35% of spontaneous abortions (Figure 1; Hassold & Hunt, 2001).

Humans are much more able to tolerate extra sex chromosomes than extra autosomes. Thus, after DS, the most common human aneuploidy is the condition known as Klinefelter's syndrome (Jacobs & Strong, 1959). Klinefelter's males have a total chromosome number of 47, which includes two X chromosomes and one Y chromosome. According to convention, these males are designated as 47,XXY individuals. Other configurations of the sex chromosomes have been observed in 47,XXX females and 47,XYY males. Compared to autosomal trisomies, these sorts of sex chromosome trisomies are fairly benign. Affected individuals generally show reduced sexual development and fertility, but they often have normal life spans, and many of their symptoms can be treated by hormone supplementation. The ability of humans to tolerate supernumerary sex chromosomes is quite remarkable, as individuals can survive with as many as four sex chromosomes. This tolerance most likely relates to both X inactivation and to the small number of genes on the Y chromosome. In fact, when cells from individuals with more than one copy of the X chromosome are analyzed under a microscope, all but one of the X chromosomes appear as condensed Barr bodies, the cytological manifestations of X-chromosome inactivation. Supernumerary copies of the Y chromosome may be tolerated because the few gene products of the Y chromosome are not required for survival.
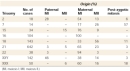
Monosomies are the opposite of trisomies, in that affected individuals are missing one chromosome, reducing their total chromosome number to 45. Cells seem to be particularly sensitive to the loss of a chromosome, because the only viable human monosomy involves the X chromosome. Females with a single copy of the X chromosome have the condition known as Turner's syndrome. Interestingly, the frequency of Turner's syndrome is significantly lower than that of sex chromosome trisomies, suggesting that a single X chromosome is insufficient for optimum cell function. The viable Turner's (45,X) females display a wide range of symptoms, which include infertility and impaired sexual development, and these individuals are usually mosaics. Most Aneuploidies Arise from Errors in Meiosis, Especially in Maternal Meiosis I
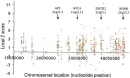
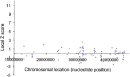
Widespread Upregulation of Chromosome 21 Genes Is Observed in Down Syndrome
An ongoing challenge for DS research is to sort through the complex set of transcriptional changes associated with DS to identify those genes whose expression is most closely linked to DS phenotypes. To this end, investigators have already constructed mouse models of chromosome 21 overexpression that reproduce some of the symptoms of DS (Antonarakis et al., 2004; FitzPatrick, 2005; Patterson & Costa, 2005). Researchers hope that these kinds of models will provide useful experimental systems for developing therapeutic interventions for this debilitating condition, which affects millions of people around the world.
References and Recommended Reading
Antonarakis, S. E., et al. Chromosome 21 and Down syndrome: From genomics to pathophysiology. Nature Reviews Genetics 5, 725–738 (2004) doi:10.1038/nrg1448 (link to article)
FitzPatrick, D. R. Transcriptional consequences of autosomal trisomy: Primary gene dosage with complex downstream effects. Trends in Genetics 21, 249–253 (2005) doi:10.1016/j.tig.2005.02.012
Hassold, T., & Hunt, P. To err (meiotically) is human: The genesis of human aneuploidy. Nature Reviews Genetics 2, 280–291 (2001) doi:10.1038/35066065 (link to article)
Jacobs, P. A., & Strong, J. A. A case of human intersexuality having a possible XXY sex-determining mechanism. Nature 183, 302–303 (1959) doi:10.1038/183302a0 (link to article)
Mao, M., et al. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics 81, 457–467 (2003) doi:10.1016/S0888-7543(03)00035-1
Patterson, D., & Costa, A. C. S. Down syndrome and genetics—A case of linked histories. Nature Reviews Genetics 6, 137–147 (2005) doi:10.1038/nrg1525 (link to article)



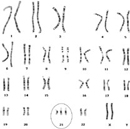
 Figure 2
Figure 2