-
March 11, 2016
Rapid RNA analysis at a single cell level: a PL... -
February 11, 2016
Free to roam: implantable optogenetic devices -
January 23, 2016
The recipe for human serotonin neurons -
January 08, 2016
Designing logical cells to be disease sensors -
December 25, 2015
The synthesis of a bacterial clock -
November 30, 2015
Folding DNA origami for chemotherapy treatments -
November 07, 2015
Gene Editing in the Natural World: Part II -
October 08, 2015
Mining the depths for new anti-biotics -
September 16, 2015
The TRAP technique gets back to basics -
August 28, 2015
A history lesson: induced pluripotent stem cells -
August 07, 2015
Let there be light: Photoactivatable CRISPR-Cas9 -
July 16, 2015
Transparent brains: mining the depths -
June 24, 2015
Restoring sight with the help of optogenetics -
June 02, 2015
Discovering new cell types one at a time -
May 12, 2015
Treating spinal cord injuries with a dual actio... -
April 20, 2015
Reducing carbon: a bacterial approach -
April 06, 2015
Propagating mutations throughout an entire popu... -
March 23, 2015
This is your brain on integrated technology -
February 28, 2015
Gene Editing in the Natural World -
February 08, 2015
Editing the Genome Quickly and Efficiently: Rou... -
January 27, 2015
Editing the Genome Quickly and Efficiently: Rou... -
January 06, 2015
Enhancing our Gut Flora for disease and allergy... -
December 19, 2014
Controlling Neurons Using Light -
June 21, 2014
Where Conscious and Subconscious Meet -
June 07, 2014
Battle of the Genetic Codes -
May 28, 2014
Blog Update -
April 08, 2014
I Attend the 55th Annual Fly Meeting! -
January 06, 2014
Going Viral for the New Year -
October 13, 2013
How to Clone Yourself, Part 2: Stem Cell Reprog... -
September 24, 2013
How to Clone Yourself, Part 1: Start Small -
September 07, 2013
What I’ve Been Doing Lately -
August 06, 2013
Is in vitro meat all it’s cracked up to be? -
July 09, 2013
Engineering in the Wild: Driving Genes into Pop... -
June 17, 2013
Bee vs. Mite! – Puerto Rico’s Honey... -
June 04, 2013
It's in the Honey! - Bee Diet and Defense Again... -
May 21, 2013
Genetic Logic Gates and Flipping DNA -
April 07, 2013
#DROS2013 Day 4 -
April 06, 2013
#DROS2013 Day 3 -
April 05, 2013
#DROS2013 Day 2 -
April 03, 2013
#DROS2013 Day 1 -
March 27, 2013
Watch for it: Tweeting and Blogging from #DROS2013 -
March 13, 2013
Undergrads Launch Genetics Education Website -
March 05, 2013
Viruses swipe bacterial immune system, turn it ... -
February 26, 2013
An Update on Artemisinin -
February 09, 2013
Editing Genomes with the Bacterial Immune System -
January 11, 2013
The One and Only Popular Synthetic Biology Book -
December 30, 2012
Beyond Genetics: DNA in Nanotechnology -
December 25, 2012
Merry Yeast Christmas! -
November 29, 2012
Engineering Viruses is Hard. Let’s Make i... -
November 02, 2012
The Oddness of California Proposition 37 -
October 26, 2012
Bird of the Week: Yellow-Bellied Sapsucker -
October 23, 2012
Sending DNA Messages Inside Viruses -
October 18, 2012
Bird of the Week: Bullfinch -
October 13, 2012
Bird of the Week: Downy Woodpecker -
October 04, 2012
My First Paper: tRNAs as Genetic Switches -
September 29, 2012
Bird of the Week: Chimney Swift -
September 26, 2012
Cheaper Genomes Should Enhance Undergraduate Ed... -
September 20, 2012
Bird of the Week: White-Breasted Nuthatch -
September 14, 2012
Science Literacy for Everyone -
September 14, 2012
Bird of the Week: Indigo Bunting -
September 11, 2012
Molecular Zoo: E. coli RNA Polymerase -
September 08, 2012
Bird of the Week: Canada Goose -
August 31, 2012
Digital Data Storage in DNA -
August 29, 2012
Bird of the Week: Double Crested Cormorant -
August 24, 2012
Modular Transcription Factors for Eukaryotic Sy... -
August 19, 2012
Bird of the Week: Barn Swallow -
August 09, 2012
Bird of the Week: Common Raven -
August 07, 2012
Molecular Zoo: The TALE’s Tale -
July 30, 2012
Artificial Beginnings: Understanding the Origin... -
July 24, 2012
What the Simulated Cell Actually Means -
July 01, 2012
Zinc Fingers: An Emerging Tool for Gene Therapy -
June 18, 2012
Your Friend the Sequence Logo -
June 07, 2012
In Living Memory: the First Steps toward Geneti... -
May 28, 2012
High-Level Control of Synthetic Systems: The Ge... -
May 07, 2012
Final Exams Beckon... -
May 02, 2012
Artemisia annua: A Vital Partner in the Global ... -
April 22, 2012
Synthetic Nucleic Acids: Beyond DNA and RNA -
April 08, 2012
Horizontal Gene Transfer: A Force for Bacterial... -
March 12, 2012
Chemicals Responsible for Bee Scouting Behavior... -
February 18, 2012
DNA Nanorobot Targets Cells for Molecular Delivery -
January 22, 2012
Seaweed Biofuel, #ArsenicLife, and Synthetic Bi... -
January 14, 2012
Soldier Bees -
January 06, 2012
Synthetic Biology is on its Way to Treating Hum... -
December 25, 2011
Computer Controlled Yeast and an E. coli LCD Sc... -
November 24, 2011
Genomes at the Thanksgiving Table -
November 06, 2011
What can Synthetic Biology Teach us About Basic... -
October 29, 2011
What can Synthetic Biology Teach us About Basic... -
October 16, 2011
What can Synthetic Biology Teach us About Basic... -
October 11, 2011
When Will a Synthetic Biologist be called to St... -
September 24, 2011
Engineering Genetic Codes -
September 10, 2011
It's Raining Caterpillars! -
September 01, 2011
Remodeling Cells with RNA -
August 18, 2011
The Promises, Demands, and Risks of Garage Biology -
August 10, 2011
DIY Hardware for the Home Biology Lab -
August 03, 2011
Synthetic Biology at Home -
July 27, 2011
The Most Interesting Paper I've Ever Read -
July 20, 2011
DNA Synthesis -
July 13, 2011
High Throughput Sequencing and Cost Trends -
July 06, 2011
Sanger Sequencing -
June 29, 2011
Thinking Like Engineers -
June 23, 2011
Artemisinin: A Synthetic Biology Success Story -
June 14, 2011
Synthetic Biology in the Spotlight -
June 06, 2011
PCR: A Revolutionary Invention -
May 31, 2011
E. coli and Chassis -
May 23, 2011
Molecular Modeling -
May 16, 2011
Student Research -
May 09, 2011
Modeling and E. coli that Count -
May 02, 2011
DNA Origami -
April 25, 2011
The Repressilator -
April 18, 2011
Abstraction -
April 11, 2011
More on Operons -
April 03, 2011
Promoters
« Prev « Prev Next » Next »




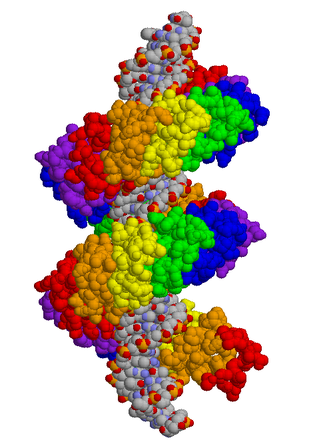 This post represents
the first in what I hope will be an ongoing series on the molecules that make
biology, well, interesting! (It also happens to be my 50th post ever on this
blog). Being alive requires taking chemistry to the extreme. At the heart of
every cell is the massive information storage molecule, DNA. DNA itself
contains the chemical code to produce all the proteins and RNAs of the cell. The
human genome alone contains the instructions for building tens of thousands of
proteins, which carry out a wide range of jobs that include breaking down food,
maintaining cell shape, switching genes on and off, and more.
This post represents
the first in what I hope will be an ongoing series on the molecules that make
biology, well, interesting! (It also happens to be my 50th post ever on this
blog). Being alive requires taking chemistry to the extreme. At the heart of
every cell is the massive information storage molecule, DNA. DNA itself
contains the chemical code to produce all the proteins and RNAs of the cell. The
human genome alone contains the instructions for building tens of thousands of
proteins, which carry out a wide range of jobs that include breaking down food,
maintaining cell shape, switching genes on and off, and more. Given its structure, biologists were quick to hypothesize
that TALEs operate by a code. Since there is a one-to-one correspondence
between protein unit and DNA base, perhaps there is also a chemical code where
particular sets of recognition amino acids in the TALE correspond to each of
the four bases of DNA. It’s since been discovered that in each repeating unit
two amino acids act to recognize a base of DNA. Since these two amino acids are
repeated in the same position of each of the TALE’s units, though their
identities vary, they are called repeat-variable diresidues (RVDs). There do
appear to be general rules, but there are a lot of factors to consider. A
recent report (Streubel et al.,
2010), for instance, recommends incorporating RVDs that bind strongly to their
recognition bases with hydrogen bonds. Some amino acids do not form hydrogen
bonds, so even though they might confer high specificity of binding, the TALE falls
off the DNA too easily. The same study showed that TALE units are not
completely modular. Asparagine + isoleucine usually binds to adenine, but not
if it is used in a repeat. Likewise with asparagine + glycine, which usually
binds thymine. However, repeats of histidine + aspartic acid bound repeats of
cytosine; and repeats of asparagine + asparagines bound repeats of guanine (but
not adenine, which they were thought to do). The original (“naïve”) TALE code
is shown in the table.
Given its structure, biologists were quick to hypothesize
that TALEs operate by a code. Since there is a one-to-one correspondence
between protein unit and DNA base, perhaps there is also a chemical code where
particular sets of recognition amino acids in the TALE correspond to each of
the four bases of DNA. It’s since been discovered that in each repeating unit
two amino acids act to recognize a base of DNA. Since these two amino acids are
repeated in the same position of each of the TALE’s units, though their
identities vary, they are called repeat-variable diresidues (RVDs). There do
appear to be general rules, but there are a lot of factors to consider. A
recent report (Streubel et al.,
2010), for instance, recommends incorporating RVDs that bind strongly to their
recognition bases with hydrogen bonds. Some amino acids do not form hydrogen
bonds, so even though they might confer high specificity of binding, the TALE falls
off the DNA too easily. The same study showed that TALE units are not
completely modular. Asparagine + isoleucine usually binds to adenine, but not
if it is used in a repeat. Likewise with asparagine + glycine, which usually
binds thymine. However, repeats of histidine + aspartic acid bound repeats of
cytosine; and repeats of asparagine + asparagines bound repeats of guanine (but
not adenine, which they were thought to do). The original (“naïve”) TALE code
is shown in the table. Though challenges remain, TALEs are interesting proteins to
engineer. They have the rare advantage of a general set of rules and a clear
starting point.
Though challenges remain, TALEs are interesting proteins to
engineer. They have the rare advantage of a general set of rules and a clear
starting point.