-
March 11, 2016
Rapid RNA analysis at a single cell level: a PL... -
February 11, 2016
Free to roam: implantable optogenetic devices -
January 23, 2016
The recipe for human serotonin neurons -
January 08, 2016
Designing logical cells to be disease sensors -
December 25, 2015
The synthesis of a bacterial clock -
November 30, 2015
Folding DNA origami for chemotherapy treatments -
November 07, 2015
Gene Editing in the Natural World: Part II -
October 08, 2015
Mining the depths for new anti-biotics -
September 16, 2015
The TRAP technique gets back to basics -
August 28, 2015
A history lesson: induced pluripotent stem cells -
August 07, 2015
Let there be light: Photoactivatable CRISPR-Cas9 -
July 16, 2015
Transparent brains: mining the depths -
June 24, 2015
Restoring sight with the help of optogenetics -
June 02, 2015
Discovering new cell types one at a time -
May 12, 2015
Treating spinal cord injuries with a dual actio... -
April 20, 2015
Reducing carbon: a bacterial approach -
April 06, 2015
Propagating mutations throughout an entire popu... -
March 23, 2015
This is your brain on integrated technology -
February 28, 2015
Gene Editing in the Natural World -
February 08, 2015
Editing the Genome Quickly and Efficiently: Rou... -
January 27, 2015
Editing the Genome Quickly and Efficiently: Rou... -
January 06, 2015
Enhancing our Gut Flora for disease and allergy... -
December 19, 2014
Controlling Neurons Using Light -
June 21, 2014
Where Conscious and Subconscious Meet -
June 07, 2014
Battle of the Genetic Codes -
May 28, 2014
Blog Update -
April 08, 2014
I Attend the 55th Annual Fly Meeting! -
January 06, 2014
Going Viral for the New Year -
October 13, 2013
How to Clone Yourself, Part 2: Stem Cell Reprog... -
September 24, 2013
How to Clone Yourself, Part 1: Start Small -
September 07, 2013
What I’ve Been Doing Lately -
August 06, 2013
Is in vitro meat all it’s cracked up to be? -
July 09, 2013
Engineering in the Wild: Driving Genes into Pop... -
June 17, 2013
Bee vs. Mite! – Puerto Rico’s Honey... -
June 04, 2013
It's in the Honey! - Bee Diet and Defense Again... -
May 21, 2013
Genetic Logic Gates and Flipping DNA -
April 07, 2013
#DROS2013 Day 4 -
April 06, 2013
#DROS2013 Day 3 -
April 05, 2013
#DROS2013 Day 2 -
April 03, 2013
#DROS2013 Day 1 -
March 27, 2013
Watch for it: Tweeting and Blogging from #DROS2013 -
March 13, 2013
Undergrads Launch Genetics Education Website -
March 05, 2013
Viruses swipe bacterial immune system, turn it ... -
February 26, 2013
An Update on Artemisinin -
February 09, 2013
Editing Genomes with the Bacterial Immune System -
January 11, 2013
The One and Only Popular Synthetic Biology Book -
December 30, 2012
Beyond Genetics: DNA in Nanotechnology -
December 25, 2012
Merry Yeast Christmas! -
November 29, 2012
Engineering Viruses is Hard. Let’s Make i... -
November 02, 2012
The Oddness of California Proposition 37 -
October 26, 2012
Bird of the Week: Yellow-Bellied Sapsucker -
October 23, 2012
Sending DNA Messages Inside Viruses -
October 18, 2012
Bird of the Week: Bullfinch -
October 13, 2012
Bird of the Week: Downy Woodpecker -
October 04, 2012
My First Paper: tRNAs as Genetic Switches -
September 29, 2012
Bird of the Week: Chimney Swift -
September 26, 2012
Cheaper Genomes Should Enhance Undergraduate Ed... -
September 20, 2012
Bird of the Week: White-Breasted Nuthatch -
September 14, 2012
Science Literacy for Everyone -
September 14, 2012
Bird of the Week: Indigo Bunting -
September 11, 2012
Molecular Zoo: E. coli RNA Polymerase -
September 08, 2012
Bird of the Week: Canada Goose -
August 31, 2012
Digital Data Storage in DNA -
August 29, 2012
Bird of the Week: Double Crested Cormorant -
August 24, 2012
Modular Transcription Factors for Eukaryotic Sy... -
August 19, 2012
Bird of the Week: Barn Swallow -
August 09, 2012
Bird of the Week: Common Raven -
August 07, 2012
Molecular Zoo: The TALE’s Tale -
July 30, 2012
Artificial Beginnings: Understanding the Origin... -
July 24, 2012
What the Simulated Cell Actually Means -
July 01, 2012
Zinc Fingers: An Emerging Tool for Gene Therapy -
June 18, 2012
Your Friend the Sequence Logo -
June 07, 2012
In Living Memory: the First Steps toward Geneti... -
May 28, 2012
High-Level Control of Synthetic Systems: The Ge... -
May 07, 2012
Final Exams Beckon... -
May 02, 2012
Artemisia annua: A Vital Partner in the Global ... -
April 22, 2012
Synthetic Nucleic Acids: Beyond DNA and RNA -
April 08, 2012
Horizontal Gene Transfer: A Force for Bacterial... -
March 12, 2012
Chemicals Responsible for Bee Scouting Behavior... -
February 18, 2012
DNA Nanorobot Targets Cells for Molecular Delivery -
January 22, 2012
Seaweed Biofuel, #ArsenicLife, and Synthetic Bi... -
January 14, 2012
Soldier Bees -
January 06, 2012
Synthetic Biology is on its Way to Treating Hum... -
December 25, 2011
Computer Controlled Yeast and an E. coli LCD Sc... -
November 24, 2011
Genomes at the Thanksgiving Table -
November 06, 2011
What can Synthetic Biology Teach us About Basic... -
October 29, 2011
What can Synthetic Biology Teach us About Basic... -
October 16, 2011
What can Synthetic Biology Teach us About Basic... -
October 11, 2011
When Will a Synthetic Biologist be called to St... -
September 24, 2011
Engineering Genetic Codes -
September 10, 2011
It's Raining Caterpillars! -
September 01, 2011
Remodeling Cells with RNA -
August 18, 2011
The Promises, Demands, and Risks of Garage Biology -
August 10, 2011
DIY Hardware for the Home Biology Lab -
August 03, 2011
Synthetic Biology at Home -
July 27, 2011
The Most Interesting Paper I've Ever Read -
July 20, 2011
DNA Synthesis -
July 13, 2011
High Throughput Sequencing and Cost Trends -
July 06, 2011
Sanger Sequencing -
June 29, 2011
Thinking Like Engineers -
June 23, 2011
Artemisinin: A Synthetic Biology Success Story -
June 14, 2011
Synthetic Biology in the Spotlight -
June 06, 2011
PCR: A Revolutionary Invention -
May 31, 2011
E. coli and Chassis -
May 23, 2011
Molecular Modeling -
May 16, 2011
Student Research -
May 09, 2011
Modeling and E. coli that Count -
May 02, 2011
DNA Origami -
April 25, 2011
The Repressilator -
April 18, 2011
Abstraction -
April 11, 2011
More on Operons -
April 03, 2011
Promoters
« Prev « Prev Next » Next »




 The
Origin of Life on Earth was certainly, in retrospect, and from the human
vantage point, the most fateful event in the history of the Universe. On a
young, tepid Earth chemistry sprung into biology and set course on a four
billion year journey that would eventually lead to us. However, all traces of the
first, primitive organisms have vanished. They were outcompeted and devoured by
their evolutionary descendents, leaving nothing to form fossils. Though we will
never be able to set eyes on the first Earthlings, the first pioneers, we can understand
what they must have been like through more subtle, indirect approaches.
Comparative biochemistry across the whole of life takes us back quite a ways,
though not to the first cells. The most recent common ancestor shared by all
living organisms—bacteria, plants, animals, fungi, archaea, and unicellular
eukaryotes like amoebae—was born long after the first cell ceased to exist. The
only way we can truly understand what life must have been like in its earliest
days is to create it ourselves.
The
Origin of Life on Earth was certainly, in retrospect, and from the human
vantage point, the most fateful event in the history of the Universe. On a
young, tepid Earth chemistry sprung into biology and set course on a four
billion year journey that would eventually lead to us. However, all traces of the
first, primitive organisms have vanished. They were outcompeted and devoured by
their evolutionary descendents, leaving nothing to form fossils. Though we will
never be able to set eyes on the first Earthlings, the first pioneers, we can understand
what they must have been like through more subtle, indirect approaches.
Comparative biochemistry across the whole of life takes us back quite a ways,
though not to the first cells. The most recent common ancestor shared by all
living organisms—bacteria, plants, animals, fungi, archaea, and unicellular
eukaryotes like amoebae—was born long after the first cell ceased to exist. The
only way we can truly understand what life must have been like in its earliest
days is to create it ourselves. Immediately
we bump up into the question “What is life?” What are we creating, exactly? Scientists
have been asking the question “What is Life?”1 for generations, and
no one has yet come up with a satisfying answer. The common answers usually
involve a combination of inheritance, evolution, and metabolism. Ed Regis makes
a good case for the latter, but that of course excludes viruses, a judgment
that I find unsatisfying3. The Nobel Laureate Jack Szostak, on the
other hand, holds that the business of defining life in such terms is pointless
and not even useful, and that what we should be focusing on is how chemistry
can transition into biology4. I’ll leave it at that.
Immediately
we bump up into the question “What is life?” What are we creating, exactly? Scientists
have been asking the question “What is Life?”1 for generations, and
no one has yet come up with a satisfying answer. The common answers usually
involve a combination of inheritance, evolution, and metabolism. Ed Regis makes
a good case for the latter, but that of course excludes viruses, a judgment
that I find unsatisfying3. The Nobel Laureate Jack Szostak, on the
other hand, holds that the business of defining life in such terms is pointless
and not even useful, and that what we should be focusing on is how chemistry
can transition into biology4. I’ll leave it at that. Creating
life, whatever we decide it is exactly, will require substantial investment and
a coherent toolset. It’s not enough to be really good at making lipid membranes
on one day and RNAs on the next; everything must come together for the simple
cell—a protocell, as they’re called—to work. I’ll be the first to admit that
the field I’m most familiar with, synthetic biology, is largely devoid of the
tools for such an undertaking. The unifying tool of synthetic biology is
synthetic DNA, whether it’s used to build genetic circuits or entire genomes.
Synthetic biology is not concerned with re-creating the Origin of Life. Despite
what was claimed by the popular press, Craig Venter’s synthetic cell2
was no Second Genesis. Converting the DNA sequence of a bacterial genome in an
online database to an actual chemical genome, and using it to bring to life a
dead (genome-free) cell was a remarkable accomplishment that will have
significant applications yet to come. But building a protocell requires
starting from basic materials, not drawing on the genetic repositories of
existing organisms.
Creating
life, whatever we decide it is exactly, will require substantial investment and
a coherent toolset. It’s not enough to be really good at making lipid membranes
on one day and RNAs on the next; everything must come together for the simple
cell—a protocell, as they’re called—to work. I’ll be the first to admit that
the field I’m most familiar with, synthetic biology, is largely devoid of the
tools for such an undertaking. The unifying tool of synthetic biology is
synthetic DNA, whether it’s used to build genetic circuits or entire genomes.
Synthetic biology is not concerned with re-creating the Origin of Life. Despite
what was claimed by the popular press, Craig Venter’s synthetic cell2
was no Second Genesis. Converting the DNA sequence of a bacterial genome in an
online database to an actual chemical genome, and using it to bring to life a
dead (genome-free) cell was a remarkable accomplishment that will have
significant applications yet to come. But building a protocell requires
starting from basic materials, not drawing on the genetic repositories of
existing organisms.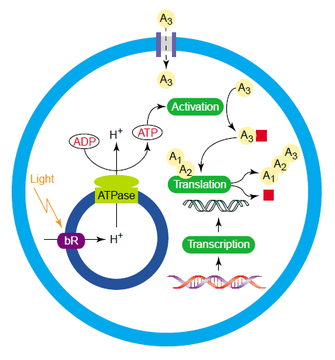 The
beauty of the protocell idea lies in its adaptability. Even if the single
replicating RNA approach fails, there are multitudes of other approaches along
a continuum of complexity. The more complex the starting point, the less true
we are to modeling the origin of life, but practical utility increases. An
intriguing proposal for a protocell microfactory is laid out in the side
figure. Protocells have the potential to drastically change our view of
manufacturing, especially when it comes to pharmaceuticals. It’s extremely
difficult to introduce a new gene pathway into an organism (say, yeast) and
have the reasonable level of efficiency required to make the endeavor
worthwhile. The cell environment is so complex that we cannot anticipate the
emergent behaviors that will crop up when a new system is inserted. However, a
protocell is far more complex, and human engineers design every component. The
schematic in the figure shows a small protocell factory that produces its own
energy from light. Light strikes a protein bound to a small internal vesicle
(that is, a vesicle inside the protocell itself), which causes it to pump
hydrogen ions inside. The excess of hydrogen ions turns the massive ATPase
protein like a water wheel as they escape, forming ATP. This universal energy
source can then drive the chemical reactions required to manufacture the
desired product. Plus, since the system is so simple it is much easier to tune
the levels of expression of the genes in the pathway required to make the
product, compared to a yeast or bacterial cell. Anything like this is still
years away, but the fact that we can imagine such a system is intriguing.
The
beauty of the protocell idea lies in its adaptability. Even if the single
replicating RNA approach fails, there are multitudes of other approaches along
a continuum of complexity. The more complex the starting point, the less true
we are to modeling the origin of life, but practical utility increases. An
intriguing proposal for a protocell microfactory is laid out in the side
figure. Protocells have the potential to drastically change our view of
manufacturing, especially when it comes to pharmaceuticals. It’s extremely
difficult to introduce a new gene pathway into an organism (say, yeast) and
have the reasonable level of efficiency required to make the endeavor
worthwhile. The cell environment is so complex that we cannot anticipate the
emergent behaviors that will crop up when a new system is inserted. However, a
protocell is far more complex, and human engineers design every component. The
schematic in the figure shows a small protocell factory that produces its own
energy from light. Light strikes a protein bound to a small internal vesicle
(that is, a vesicle inside the protocell itself), which causes it to pump
hydrogen ions inside. The excess of hydrogen ions turns the massive ATPase
protein like a water wheel as they escape, forming ATP. This universal energy
source can then drive the chemical reactions required to manufacture the
desired product. Plus, since the system is so simple it is much easier to tune
the levels of expression of the genes in the pathway required to make the
product, compared to a yeast or bacterial cell. Anything like this is still
years away, but the fact that we can imagine such a system is intriguing.

















["The Emergence and Early Evolution of Biological Carbon-Fixation, Braakman et al, PLoS Comp Bio 2012; "Ancestral lipid biosynthesis and early membrane evolution", Peretó et al, TRENDS in Bio Sci 2004.]
A persistent metabolic core then. But if early stromatolites were indeed photophilic, an early anoxic photosynthesis may have evolved.
It would have liberated cells from local redox sources like hydrothermal vents without much specialization. It is relatively easy to evolve for both protection and energy source (many compounds works), and the low level of light required would fit deep habitats with surface UV (no ozone).
While it is true that comparative methods not take us back to the first cells, I note that they have penetrated deep. Protein fold family work has covered the DNA UCA as well as the RNA/protein world before it.
By a fold clock proxy the RNA/protein world was ~ 20 % of time, the DNA UCA period ~ 20 %, and the diversification into the domains of modern cells the rest. So much of the ancestral 50 % of cellular history is still covered. (But at increasingly lower resolution of course.)
["The evolution and functional repertoire of translation proteins following the origin of life", Goldman et al, Biol Dir 2010; and similar works.]