Abstract
To explain anaerobic nitrite/nitrate production at the expense of ammonium mediated by manganese oxide (Mn(IV)) in sediment, nitrate and manganese respirations were investigated in a strain (Shewanella algae C6G3) presenting these features. In contrast to S. oneidensis MR-1, a biotic transitory nitrite accumulation at the expense of ammonium was observed in S. algae during anaerobic growth with Mn(IV) under condition of limiting electron acceptor, concomitantly, with a higher electron donor stoichiometry than expected. This low and reproducible transitory accumulation is the result of production and consumption since the strain is able to dissimilative reduce nitrate into ammonium. Nitrite production in Mn(IV) condition is strengthened by comparative expression of the nitrate/nitrite reductase genes (napA, nrfA, nrfA-2), and rates of the nitrate/nitrite reductase activities under Mn(IV), nitrate or fumarate conditions. Compared with S. oneidensis MR-1, S. algae contains additional genes that encode nitrate and nitrite reductases (napA-α and nrfA-2) and an Outer Membrane Cytochrome (OMC)(mtrH). Different patterns of expression of the OMC genes (omcA, mtrF, mtrH and mtrC) were observed depending on the electron acceptor and growth phase. Only gene mtrF-2 (SO1659 homolog) was specifically expressed under the Mn(IV) condition. Nitrate and Mn(IV) respirations seem connected at the physiological and transcriptional levels.
Similar content being viewed by others
Introduction
In a large part of the sediment floor, organisms use terminal electron-accepting molecules, alternatives to oxygen for energy generation1. The most abundant molecules comprise NO3−, Mn(IV), Fe(III), SO42− or organic matter resulting from fermentation. In sedimentary systems, their repartition is often stratified on a permanent or seasonal basis. Just below the oxic zone, the suboxic zone is characterized by high concentrations of oxidized inorganic compounds, such as nitrate, manganese oxides (Mn(IV)) and iron oxy/hydroxides (Fe(III)) that are in close proximity1. Nitrate is generally known to be generated in aerobiosis by nitrification from ammonium, which results from the mineralization of organic matter. Iron and manganese are commonly encountered in the Earth’s crust. Water percolating through soil and rock can dissolve minerals containing iron and manganese and hold them in solution. After oxidation, mainly by water-soluble oxygen, the metal oxides precipitate and settle into the sediment.
Biogenic redox reactions have been recognized as paramount for global biogeochemical cycles. From the geochemical point of view, manganese-oxides are commonly described as powerful oxidants capable of modifying or creating new pathways of oxidation in the N cycle2,3,4,5,6,7,8,9. Luther et al., (1997) have shown strong links between Mn and N cycles and have proposed several putative reactions causing short-cuts in the N cycle6. Biogeochemical evidence has been found for such reactions, for example, the enhanced N2 production in the presence of Fe(III) or Mn(IV) in marine sediment samples10. Furthermore, laboratory incubation experiments with Atlantic coastal sediment showed a significant increase in N2O production upon the addition of Mn(IV)11. The same trend was observed in mangrove sediment from India: N2O production was higher in sediment subjected to ferromanganese mining operations in comparison to a pristine site12. Among NH4+ oxidation processes, the role of Mn(IV) compounds in alternative anaerobic metabolism has often been suggested to explain the presence of unexpected bursts of nitrite/nitrate in deep layers of sediment2.
However, due to the complex recycling of redox species, these processes are difficult to discern and therefore are poorly understood. Javanaud et al., (2011) isolated a denitrifying strain belonging to the genus Marinobacter from Atlantic coastal sediments and used it to demonstrate N2 production from NH4+ 13. Anaerobic nitrification–denitrification in the presence of manganese oxides was suggested, and a putative formation of nitrate or nitrite at the expense of ammonium by this manganese oxide reducing strain was proposed13.
Shewanella oneidensis MR-1 is the most-described and well-known strain capable of dissimilatory metabolism of manganese oxides14. The genus Shewanella is known for its versatile electron-accepting capacity, which allows the coupling of the decomposition of organic matter to the reduction of the various terminal electron acceptors that these organisms encounter in their stratified environments. The model species Shewanella oneidensis MR-1 presents the notable properties of using more than 10 terminal electron acceptors and of transferring electrons to solid metal oxides. Currently, bacteria of the genus Shewanella comprise more than 50 species that inhabit a wide range of marine environments and ecosystems. In the frame of the study of anaerobic ammonium oxidation mediated by Mn(IV), a pool of Shewanella strains able to perform nitrate and Mn(IV) reductions was isolated from the Atlantic coastal sediment13. The genome of one of these strains, Shewanella algae C6G3, has been recently sequenced and differences in the gene organization compared with Shewanella oneidensis MR-1 were identified (Fig. 1A,B).
(A) Presentation of the potential operons carrying genes of NAP systems (dissimilatory nitrate reduction, DNR), NRF systems (dissimilatory nitrite reduction into ammonium, DNRA) and Mtr systems (reduction of external electron acceptors). The presented genes correspond to the organization of S. algae C6G3 genome check manually (BLASTP against S. algae strain ACDC and JCM 21037 on JGI IMG/ER, nrfA-2 = SA002_01180, mtrH = SA002_00064 and mtrF-2 = SA002_00782 and mtrEFC), or automatically annotated (the other genes). (B) Presentation of corresponding operons carrying the genes of interest in S. oneidensis MR-1. Note that nrfA-2, napEDA-1B-1C, nrfBC and mtrH are absent in the S. oneidensis MR-1 genome (bold).
To complement information obtained from the literature on gene-encoding proteins involved in Mn(IV) respiration based on mutagenesis in S. oneidensis MR-115,16,17, our study explores the kinetics of the expression level by qRT-PCR of the gene-encoding potential “end effectors” for Mn(IV) (omcA, mtrC, mtrF, mtrH) and nitrate/nitrite (napA, nrfA) respirations, two oxidized inorganic compounds in close vicinity in sedimentary systems during the growth of S. algae C6G3. Two other genes, SA002_00782 and SA002_01180, which encode a putative decaheme c-type cytochrome MtrF and a putative cytochrome c-552 precursor, respectively, were also monitored. During growth with Mn(IV) or nitrate as the electron acceptor, the kinetics of nitrate, nitrite, Mn(II) production or consumption were investigated to obtain system-level insights on anaerobic NO2/3− production in the presence of Mn(IV). Moreover, the gene expression levels were compared under the two respiration conditions, growth with fumarate being used as control. Our results demonstrate a transitory nitrite accumulation during growth with Mn(IV) in agreement with the concomitant expression of nitrate/nitrite reductase genes and nitrate and nitrite reductase activities. Concerning the OMC, their expression is growth-phase dependent and peaked at the end of the exponential phase under the nitrate and Mn(IV) conditions in contrast to that observed for the control with fumarate. Except for mtrH, all OMC tested genes presented higher expressions in nitrate or Mn(IV) compared with the control (fumarate). However, only the new gene mtrF-2 was specifically expressed during the entire growth period in Mn(IV) compared with nitrate and fumarate conditions.
Results and Discussion
Nitrate and Mn(IV) respiratory gene analysis, classical and “new genes” identification
From the sequence of the genome of S. algae C6G3 (GenBank ID JPMA00000000 (JPMA01000001-JPMA01000043)18) the genes encoding NAP, NRF and Mtr systems giving electrons to nitrate, nitrite or ion metal oxide, respectively and ensuring the electron transfer from the cytoplasm to the periplasm and extracellular compounds was proposed for this strain (Fig. 1A). Except for S. oneidensis MR-1 and S. denitrificans OS217, the genome of the majority of the species belonging to the genus Shewanella including S. algae C6G3 and S. algae ACDC present two isoforms of the NAP operon, Nap-α and Nap-β19,20 insuring the reduction of nitrate into nitrite by a periplasmic nitrate reductase (NapA) (Fig. 1A,B).
The nitrite reduction step into ammonium was performed by a periplasmic cytochrome c nitrite reductase NrfA (Fig. 1A,B)21. In addition to the NAP-α operon, a gene potentially involved in the transformation of the nitrogenous element was identified in the genome of Shewanella algae C6G3 (formally SA002_01180) and annotated as formate-dependent nitrite reductase, periplasmic cytochrome c‐552 subunit with KO:K03385 (EC 1.7.2.2) and COG3303 according to the JGI IMG/ER platform. After sequencing and verification of the sequence integrity, this gene was herein referenced as nrfA-2. The BLASTp of the sequence on NCBI revealed the presence of nrfA-2 homologs among the sequenced genomes only in the genus Shewanella and Ferrimonas with an occurrence of 61% (n = 29) and 100% (n = 4), respectively (see Fig. S1 in the supplemental material). As for the Nap-α operon, nrfA-2 is absent in the S. oneidensis MR-1 genome (Fig. 1B). The role of the cytochrome c-552 is unknown. The study of Clark et al., (2013)22 on S. algae ACDC excluded its involvement in nitrite reductase activity because the presence of the nrfA-2 homolog in the S. algae ACDC genome did not permit the nitrite reduction activity, when the nrfA gene is inactivated by the presence of a transposon22. Other authors have also noted the absence of any role of this gene in the respiration of the nitrate or nitrite in S. piezotolerans WP320.
The respiratory external electron acceptor machinery (mainly Fe(III) and Mn(IV)) has been extensively studied in the model strain S. oneidensis MR-1. These studies have identified the effectors of the electron transport chain from the cytoplasm to the cell surface where the external electron acceptor is reduced via the Mtr system23. The Mtr system consists of a group of transmembrane and periplasmic cytochrome encoded genes organized in an operon24 comprising from the cytoplasm to the outer membrane: (I) the periplasmic tetraheme cytochrome c CymA20,25,26,27, (II) the cytochromes MtrAB and their homologues MtrDE and (III) the extracellular deca-haem c-type cytochromes of the OmcA/MtrC family28 (OMC) responsible for the reduction of extracellular metal oxides and flavins29,30,31,32.
In S. algae C6G3, the Mtr operon is composed of 8 genes mtrDEF-omcA-mtrH-mtrCAB (Fig. 1A). The mtrH gene is absent in S. oneidensis MR-1 Mtr operon (Fig. 1B). This extracellular deca-haem c-type outer-membrane -cytochrome24,33,34 is found in only 23% of the Shewanella strains tested (n = 29, data not shown), and it is referenced in S. halifaxensis HAW- EB4 (Shal_2783) and S. piezotolerans WP3 (swp_3277)33,34. Extensive studies on the genome of S. oneidensis MR-1 have also revealed the presence of 42 potential c-type cytochromes (soluble periplasmic proteins, cytoplasmic membrane proteins and outer membrane lipoproteins) distributed on its genome35. Two out of the 5 identified as outer membrane lipoproteins are external to the Mtr system: SO1659 (decaheme c) and SO2931 (diheme c). Investigations into the genome of S. algae C6G3 have identified only the homolog of SO1659 as SA002_00782. This gene has been annotated as a decaheme c-type cytochrome, OmcA/MtrC family in the JGI IMG/ER platform and as mtrF in the RAST platform; thus, it was herein referenced as mtrF-2 to distinguish it from mtrF of the Mtr operon. The gene mtrF-2 is distributed in 100% of the tested strains belonging to the Shewanella genus (n = 29; see Fig. S2 in the supplemental material); this ubiquity suggests a paramount role.
Connection between NO3− and Mn(IV) respiration on the physiological level through nitrite production associated with Mn(IV) respiration
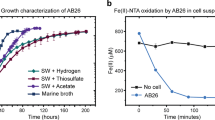
Shewanella algae C6G3 is capable of using nitrate or Mn(IV) as the sole electron acceptor under anaerobic conditions18, with a generation time 2-fold shorter (0, 64 h vs. 1, 32 h), and a culture yield 25-fold higher for the nitrate condition (Fig. 2A,B).
For the growth of S. algae C6G3 with nitrate, there was a rapid consumption of nitrate correlated with nitrite accumulation which was rapidly consumed (Fig. 2A), the same trend was observed for S. algae ATCC 51192 (see Fig. S3A in the supplemental material). For S. algae C6G3 growth on Mn(IV), Mn2+ production is observed to reach approximately 70% of the initial concentration of Mn(IV). In contrast to culture of S. oneidensis MR-1 (Fig. 2C), there is a low but reproducible nitrite accumulation (reaching ~2 μM) during the exponential phase of growth for the S. algae strains C6G3 and ATCC 51192 (Fig. 2B; see Fig. S3B in the supplemental material). During the stationary phase of growth, the nitrite concentration is maintained then decreases from 18 h to 40 h with a concomitant 3-fold increase in the number of 16 S rDNA (1.63 × 109 genes/mL at 18 h; 6.2 × 109 genes/mL at 40 h) (Fig. 2B). Neither nitrate nor nitrous oxide was detected in the cultures of S. algae in the presence of Mn(IV). The nitrite burst was not observed in media without cells (data not shown) or in cultures with Fe(III) as the electron acceptor (Fig. 2D for 1 mM Fe(III), data not shown for 3 mM Fe(III)).
The quantity of S algae C6G3 cells generated during growth under manganese oxide reducing conditions and in minimal medium with lactate as the sole carbon and energy source, was proportional to initial Mn(IV) concentration in the range 1–3 mM (Fig. 3) whereas a plateau value was observed for 6 mM of Mn(IV) probably due to an inhibitory effect. Reduction of Mn(IV) changed the color of the precipitates from dark brown to whitish rosy-red. For initial Mn(IV) concentration of 3 or 6 mM, soluble Mn(II) quantified at the end of the growth never exceeded 1.7 mM indicating that most Mn(II) was associated with the solid phase. The mineral formed was presumably MnCO3.
In the culture with 3 mM of Mn(IV), the oxidation of 5.4 mM of lactate into acetate (4.15 mM) and CO2 requires at least the reduction of 10 mM of Mn(IV) to Mn(II).

The culture experienced electron acceptor limitation, since only 3 mM Mn(IV) were available.
In consequence, under these conditions we can assume that another electron acceptor was involved in lactate oxidation. The small amount of nitrite (2 μM) accumulated in media containing NH4Cl as the nitrogen source during manganese reduction suggested the possibility that anaerobic ammonium oxidation into nitrite or nitrate occurred and their subsequent utilization as electron acceptors and reduction into ammonium.
Investigations of the impact of the Mn(IV) concentration on nitrite production by cultures of S. algae C6G3 have been also performed in a range comprised between 0.05 to 6 mM (Fig. 3). A nitrite accumulation has been observed for the initial concentrations of the metal oxide above 0.6 mM, whereas no nitrite could be detected for 0.05 and 0.2 mM. No correlation between the nitrite accumulation and the Mn(IV) initial concentration was observed (n = 15, p < 0.05, Fig. 3)). Due to its rapid production and consumption, nitrite could be a reactive short-lived intermediate and its level of accumulation depends only on the difference in the rates of production and consumption.
Several studies have highlighted nitrate and nitrite production in sediment slurries incubated under anaerobic conditions upon the addition of Mn(IV)13,36, suggesting a putative production of nitrate for at least some strains. Taking into account the above data, the nitrite production in culture of S. algae strains C6G3 and ATCC 51192, thus seems likely to be of biotic origin. The nitrite concentration (~2 μM) observed in a pure culture of S. algae strains C6G3 and ATCC 51192 is in the same range as that observed in Mn(III/IV)-rich marine sediment2 (~5 μM of NO2/3−) or in an enriched mesocosm36 (3.3 to 4.9 μM of NO2/3−). Furthermore, as observed in our study, several authors have also noted the transient nature of this accumulation in sediment13,36. However, the Mn(III/IV) concentrations necessary to obtain in vitro NO2/3− peaks are higher than the values found in Mn(III/IV)-rich marine sediments where this phenomenon has been observed (e.g., ~0.2 mM of Mn(III/IV)2).
Two hypotheses for the tight coupling of the nitrogen and manganese cycles have been proposed (Fig. 4). The first is the direct oxidation of NH4+ to NO2/3− via the reduction of manganese oxides (Equation 2 and pathway 1 Fig. 4) as proposed by several authors2,4,6,8 from results obtained from anoxic sediment.
The first one (1) implies a direct biotic oxidation of NH4+ by Mn(IV), whereas the second one (2 ABC) involves an abiotic generation of Mn(III) via the comproportionation reaction between Mn(IV) and Mn(II) (2A), followed by an abiotic oxidation of NH4+ by this highly reactive Mn(III) into NH2OH and Mn(II) (2B), and finished by a biotic disproportionation of NH2OH into NO2− and NH4+ (2 C). Dotted arrow: abiotic reaction, full arrow: biotic reaction. Cox: oxidized electron donor, Cred reduced electron donor.

The second hypothesis involves Mn(III) which is a labile intermediate formed in the presence of Mn(IV) and Mn(II). Mn(III) is extremely unstable in solution. The available Mn(III) produced herein would catalyze NH4+ oxidation into N2 or NH2OH that would be subsequently be subjected to a biological disproportionation into NH4+ and NO2− (pathway 2 Fig. 4), in analogy with the anoxic oxidation of H2S to SO42− previously described37.
Overexpression of genes encoding for enzymes associated with dissimilatory reduction of nitrate (nap) and nitrite (nrfA) and enzymatic activity of the encoding gene under manganese oxide reducing conditions could be an indirect proof of the production of nitrate/nitrite and its subsequent reduction via dissimilatory nitrate reduction into ammonium pathway.
Expression of nitrate respiratory genes and enzymatic activity of the encoding proteins in cells grown the presence of nitrate or Mn(IV) as the electron acceptor
The rpoD-relative expression level of the genes involved in the terminal transfer of electrons to nitrate and nitrite was investigated because their gene products could be involved in the nitrite production. In S. oneidensis MR-1, the expression level of numerous genes has been tested by short incubation without growth in the presence of different electron acceptors38. In the presence of nitrate in comparison with fumarate, expression of napA and of nrfA increase of 4.9 and 20.5-fold, respectively, suggesting an induction of these genes by nitrate/nitrite, whereas in the presence of Mn(IV), napA and nrfA expression levels were close to that of the fumarate control (i.e., 1.98 and 1.24). Another study on S. oneidensis MR-1 has shown that napA is responsive to nitrate, whereas nrfA is responsive to both nitrate and nitrite39. S. algae C6G3 presents two isoforms of napA with 73% similarity on the nucleotide level. It was not possible to quantify their individual expression level with the chosen primer set40. In the S. algae C6G3 control cultures (fumarate), the napA and nrfA expression level relative to rpoD remained stable during growth ranging from 0.1 to 1. In the culture of S. algae C6G3 with nitrate, the expression level of the nitrate reductase genes napA (napA-α + napA-β) was induced during the exponential growth phase (~300 relative to rpoD), corresponding to the consumption of nitrate and the production of nitrite (Fig. 5A). The expression level of napA decreased rapidly during the stationary phase of growth as nitrate is consumed. The expression level of nrfA encoding the dissimilatory nitrite reductase was correlated with the curve of nitrite; it reached its maximum (~60 relative to rpoD) when nitrite peaked (bold arrow Fig. 5C) and decreased with its consumption during the stationary phase of growth. Unexpectedly, under the Mn(IV) condition, there are expression levels of these genes that are known to be characteristic of nitrate and nitrite respirations during the exponential phase of growth. At the end of the exponential phase of growth (Fig. 5B,D), the expression level of napA reached the same level as that found under the nitrate conditions (~250 relative to rpoD). For the nrfA expression level, an induction in the Mn(IV) containing cultures was also observed during the exponential phase of growth, reaching 2-fold that of the control (fumarate, see Fig. 5G), but remaining 7-fold lower than in the presence of nitrate (Fig. 5D). Induction of napA and nrfA genes in the presence of Mn(IV), and the difference in the expression level between fumarate and Mn(IV), are in agreement with a production of nitrate/nitrite that would induce the expression of napA and nrfA.
(A–G) napA, nrfA and nrfA-2 gene expression level along bacterial growth with nitrate or manganese as the electron acceptor. The time of the kinetic corresponding to the nitrite peak is marked with a bold arrow. Cultures with fumarate (G) correspond to negative control. Standard deviation of growth curves has not been materialized for readability.
The expression level of the nrfA-2 gene was also investigated. Under the nitrate condition, its overall level was lower than that of nrfA (maximal expression level for nrfA-2 and nrfA relative to rpoD reached ~6 versus ~60, respectively) but followed the same trend with a sharp increase in the exponential phase of growth and a sharp decrease in the stationary phase of growth (Fig. 5E). In the control (fumarate, Fig. 5G), the expression of this gene followed the same trend as under the nitrate condition. In contrast, in the presence of Mn(IV), the expression level of nrfA-2 was higher than that of nrfA (expression rate relative to rpoD ~15 versus ~8, respectively, for nrfA-2 and nrfA for the maximum expression level) (Fig. 5F). At the beginning and during the stationary phase of growth, the expression of nrfA-2 was 1.9 and 2.9-fold higher than under the nitrate condition, suggesting the maintenance of its role.
To verify that the Mn(IV) induction of genes involved in the N cycle (nrfA, napA) leads to the effective reduction of nitrate and nitrite by the bacteria, crude extracts of cells grown in the presence of Mn(IV) were assayed for nitrate and nitrite reductase activities. When fumarate was the terminal electron acceptor in the medium, no significant nitrate or nitrite reductase activity was detected (Table 1). As expected, when cells were grown in the presence of nitrate, both enzymatic activities were measured at an optimal level in the extract and an increase of 443 and 26 fold was observed for the nitrate and the nitrite reductase activities, respectively. Interestingly, the presence of Mn(IV) during the cell growth leads also to a higher level of nitrate and nitrite reductase activity of 150 and 5 fold for the nitrate and nitrite reductase respectively, to those measured in the crude extract of cells grown with fumarate. This result indicates that Mn(IV) induces the production of enzymes able to catalyze nitrate and nitrite reduction. Since our transcriptomic analyses show unambiguously an increase of expression-level of genes involved in nitrate and nitrite respirations, we propose that the measured activities depend on the products of these genes.
Taken together with the induction of napA and nrfA by nitrate/nitrite observed earlier in S. oneidensis MR-138, these data are in agreement with the hypothesis of an intracellular production of nitrite, and possibly nitrate, in S. algae C6G3 in the presence of Mn(IV). In the case where nitrate would be produced under the presence of the Mn(IV) condition, the strong expression level of napA and the activity of the encoding enzymes could result in the rapid reduction of nitrate, preventing its detection. In contrast, the lower and delayed expression level of nrfA in the presence of Mn(IV) could favor the nitrite accumulation observed in physiologic studies. The localization of nrfA-2 close to NAP-α on the S. algae C6G3 genome, together with its higher expression level under the Mn(IV) condition, are in agreement with a direct or indirect role in the production of nitrite under the Mn(IV) condition. Both physiologic transcriptomic and enzymatic data suggest nitrite production at the expense of ammonium in anaerobiosis and in the presence of Mn(IV). Many Bacteria, Archaea and Fungi are known to oxidize ammonium into nitrite by a pathway called nitrification, oxygen being the electron donor for the prokaryote41. Bacterial and archaeal autotrophic nitrification requires the concerted action of an ammonia monooxygenase (amo) and hydroxylamine oxidase (hao). Fungal nitrification is not an energetic pathway and involves the reaction of reduced organic compounds with hydroxyl radicals produced in the presence of hydrogen peroxide and superoxide. Our data show anaerobic ammonium oxidation in the presence of manganese oxide, whereas neither amo nor hao genes are found on the genome of S. algae, suggesting other mechanisms. Several hypotheses could be issued: (i) the presence of an unidentified new gene able to perform ammonium oxidation; (ii) the reverse reaction by an enzyme, such as NrfA or NrfA-2 for example, due to the electron flow equilibrium; and (iii) a secondary reaction generated by a putative oxidative stress.
Expression of OMC genes in the presence of nitrate or Mn(IV) as the electron acceptor
To better understand the relative importance of the outer membrane cytochrome in Mn(IV) respiration, comparative monitoring of their gene expression levels was performed between the nitrate and Mn(IV) cultures for the entire growth period (Fig. 6) with fumarate as a control. Mn(IV) reduction is performed in two steps (i) reduction of Mn(IV) to Mn(III) and (ii) reduction of Mn(III) to Mn2+, both steps seem to be dependent on the Mtr system, but only the second step causes CO2-production42.
(A–F) Outer membrane Cytochrome (OMC) gene expression levels. mtrF-2, mtrC and mtrH gene expression level along bacterial growth with nitrate or manganese as the electron acceptor (A–F). Comparison of expression of 5 OMC in nitrate, Mn(IV) and fumarate condition (negative control) (G–I). Standard deviation of growth curves has not been materialized for readability.
In the control, the expression levels of all OMC genes were relatively stable during growth, with mtrF-2 below 0.1, omcA and mtrF approximately 1, mtrC approximately 10 and mtrH approximately 600 (relative to rpoD, see Fig. 6G–I). In the presence of nitrate or Mn(IV), their expression level increased during the exponential phase of growth then decreased during the stationary phase of growth, except for mtrH and mtrC, which remained relatively stable at the end of the growth on Mn(IV). The highest expressed gene is mtrF (expression rate between 103 to 104) and its expression profiles for nitrate and Mn(IV) conditions are strictly superposed (Fig. 6G,H). Among the cytochrome genes under both conditions, mtrH has the second highest expression level after mtrF (Fig. 6). The omcA gene appeared highly expressed in nitrate compared with Mn(IV) during the entire growth period. In contrast, the mtrC and mtrH gene expression levels were higher in the Mn(IV) condition only in the early exponential phase (Fig. 6). The S. oneidensis MR-1 mutant analysis has shown that omcA is not essential for Mn(IV) respiration, whereas mtrC could be specific for the second step of Mn(IV) respiration (e.g., the reduction of Mn(III) in Mn2+)43. Furthermore, each gene has separate promoters, with omcA transcribed alone whereas mtrC is included in the mtrCAB operon44. Our results for S. algae C6G3 are consistent with these finding. The non-specificity of omcA is strengthened by having the same range of expression levels in the nitrate and Mn(IV) cultures with a slightly higher value for the nitrate (Fig. 6G,H). The putative specificity of mtrC in the Mn(IV) respiration suggested before is strengthened by our results showing that mtrC is significantly over-expressed in the early exponential phase (where Mn2+ concentration reached 500 μM) in the manganese oxide relative to the nitrate condition (Fig. 6C,D).
The gene mtrF-2 (homolog of SO1659 in S. oneidensis MR-1 genome) is significantly over-expressed in the presence of Mn(IV) versus nitrate all during growth. At its maximal expression level, it is 6-fold higher for the Mn(IV) condition (Fig. 6A,B) compared with nitrate and 5000-fold higher compared with fumarate (Fig. 6I). Furthermore, low differences between the early-exponential phase and the beginning of the stationary phase of growth of S. algae C6G3 in the Mn(IV) condition were observed. From a relative point of view, the OMC expression level profiles show a higher expression level of mtrF-2 vs. mtrC and omcA in the Mn(IV) condition whereas in the nitrate condition, mtrF-2 is less expressed than mtrC and omcA (Fig. 6G,H). The role of the OMC mtrF-2 (not belonging to the Mtr operon), is still unclear. Bucking et al., (2010) showed that SO_1659strep, a ΔOMC mutant complemented with SO1659 (mtrF-2), was unable to restore the rate of Fe(III) and Mn(IV) reduction45. Previous studies have shown no significant impact on the respiration of Mn(IV) in S. oneidensis MR-1 wild type versus ΔSO1659 (ΔmtrF-2)46,47 at the beginning and at the end of growth. However, the absence of an impact could be due to the colorimetric technique used for the Mn(IV) measures. Because the color of the compound generated is indifferent to the forms of Mn(III) to Mn(VII)48, the first step of the reduction of Mn(IV) (Mn(IV) to Mn(III)) could not be monitored. The over-expression level of mtrF-2 in the presence of Mn(IV), together with the low difference of its expression level between the early-exponential and exponential stages of growth suggest a role in the reduction of Mn(IV) and putatively in the Mn(IV) to Mn(III) reduction step.
The regulation of the OMC expression genes belonging to the Mtr operon seems complex and presents differences between them despite their good synteny, except for mtrF-2. Growth phase appeared as an important parameter for the analysis of expression levels of these genes.
Conclusion
Due to mineralization of organic matter, ammonium concentration present usually higher concentration than that of nitrate or nitrite in sediment, with and increasing tendency with deep, whereas nitrate. nitrite or Mn(IV) show peaks due to net balance between oxidative and reductive process1. Physiologic and transcriptomic analysis of S. algae C6G3 originating from marine sediment revealed new features regarding nitrate respiration and Mn(IV) and connection between manganese and nitrogen cycles.
Regarding the effectors of the manganese oxide respiration, the data from this study raised interesting points. The omcA and mtrF genes do not seem specifically involved in Mn(IV) respiration. In contrast, the mtrC gene showed a specific induction compared to the nitrate condition, and their products may be involved in the second stage of manganese oxide respiration, which is Mn(III) → Mn2+. The clearest and most surprising result of this study is the overexpression under the Mn(IV) condition, throughout the entire growth period, of the mtrF-2 gene, which does not belong to the Mtr operon. Its strong induction at the beginning and middle of the exponential phase supports the hypothesis of a specific role in the first step of the reduction of manganese oxide which is Mn(IV) → Mn(III).
Regarding connection between nitrogen and manganese cycles, results of this study showed an anaerobic nitrite production in the presence of Mn(IV), with ammonium as the sole nitrogen source. This phenomenon has been already observed in situ and in a laboratory microcosm, but this is the first time that this original process has been monitored with a single microorganism. The low nitrite concentration and the absence of nitrate detection is explained by the capacity of dissimilatory reduction of nitrate into ammonium of S. algae C6G3. The high-level expression of the napA gene in the nitrate condition (where nitrate is reduced into nitrite) and in Mn(IV) condition at the end of the exponential phase of growth indicates the quick reduction of nitrate into nitrite, that is also confirmed by enzymatic nitrate reduction data. Although the expression level of the nrfA gene in the Mn(IV) condition does not reach that detected under nitrate condition, the expression level of this gene throughout the experiment and the enzymatic data of nitrite reduction in the Mn(IV) condition could explain the low nitrite concentrations detected. All of these results provide evidence for NO2/3− production. The role of the nrfA-2 gene, or of another gene, is one hypothesis for this anaerobic production of nitrite in the presence of Mn(IV), together with the putative reverse reaction of an enzyme such as NrfA or NrfA-2, for example due to electron flow equilibrium or a possible secondary reaction generated by a putative oxidative stress.
Materials and Methods
Growth conditions
S. oneidensis MR-1 was grown anaerobically at 28 °C in a medium containing 86 mM of NaCl, 20 mM of NH4Cl, 50 mM of MgSO4, 0.43 mM of K2HPO4, 13.2 μM of FeSO4 and a trace mineral and vitamin solution49 and with Mn(IV) (1 mM) as the sole source of electron acceptors and with DL-lactate (25 mM) as a carbon and energy source.
S. algae C6G3 and S. algae ATCC 51192 were grown anaerobically at 28 °C on artificial sea water50 with DL-lactate (25 mM), K2HPO4 (0.43 mM), FeSO4 (13,2 μM) and trace mineral and vitamin solution49. This medium contains NH4+ at 20 mM. Yeast extract (0.01%) was added only in the cultures dedicated for transcriptomic analysis because it allows increasing growth yield without affecting the metabolism (data not shown). The chosen Mn(IV) concentration is a compromise between growth yield and RNA quality because Mn(IV) favor growth, but Mn2+ affects RNA integrity. Nitrate (2.5 mM), Mn(IV) (from 0.05 mM to 6 mM), iron oxide (1 or 3 mM) or fumarate (2.5 mM), in the control, was added as the sole source of electron acceptors. No detectable trace of nitrite or nitrate was observed in the Mn(IV), iron oxide or fumarate initial mediums. Manganese oxide was synthesized just before the beginning of the culture following the protocol of Laha et al., 199051 to insure its bioavailability. Iron oxide was also synthesized just before growth, following the protocol of Misawa et al., (1973)52. The media were inoculated with the aerobic pre-culture at a ratio of 1:50 in the early exponential phase (OD600nm ~0.020). Anaerobiosis was obtained with a 10-min gas flush (N2). All cultures were prepared at least in duplicate.
Kinetics of growth
Due to the opacity of the culture in the presence of the metal oxides, the abundance of Shewanella in the different cultures was determined by q-PCR. The increase of cellular abundance was monitored through the increase of genome number that has been measured according to the increase of the number of 16 S rDNA genes. On cultures with nitrate as electron acceptor, both OD600 and 16 S rDNA genes number were determined during growth. Optical density appeared to be correlated with the log of 16 S rDNA genes, with a R2 of 0.95. Real time PCR reactions (20 μL) contained 2 μL of diluted culture (1:10) in molecular biology grade water (5PRIME), 0.5 μM of each 16 S primer (see Table S1 in the supplemental material) and 10 μL of SsoAdvancedTM SYBR® Green Supermix (Biorad Lab, USA) and were performed in duplicate on CFX96 Real Time System (C1000 thermal cycler, Bio-Rad Lab, USA) with the following reaction parameters: 2 min at 98 °C, 30 amplification cycles with 5 sec at 98 °C, 10 sec at 57 °C, 20 sec at 72 °C, followed by the melting curve step (between 65 °C to 95 °C, increase of 0.5 °C with a time step of 5 sec) to check the specificity of the amplification product. The absolute quantifications were performed with a standard curve obtained from the dilutions of a DNA plasmid, harboring a ribosomic gene of known concentration as described previously53.
The nitrite and nitrate concentrations during growth were measured on an Auto-Analyzer according to Treguer and Le Corre instruction’s54. The threshold concentration for nitrate or nitrite is 0.2 μM. The dissolved manganese (Mn2+) assay was carried out by colorimetry following the protocol of Chin et al., (1992) with a threshold concentration of 1 μM55.
RNA extraction
The samplings for RNA extractions were carried out at three points of the time series: the early exponential phase, the exponential phase and at the beginning of the stationary phase of growth. Fifteen milliliters of culture were centrifuged for 15 min at 4500 rpm. Cell pellets were suspended in 350 μL of RLT buffer of RNeasy® kit (Qiagen) containing 3.5 μL (1%) of 2-Mercaptoethanol (Aldrich) and were frozen in liquid nitrogen and then conserved at −80 °C until extraction. Then, the total RNA was purified using RNeasy® Mini QIAcube Kit (Qiagen) using the protocol “Purification of total RNA from animal tissues and cells” (eluted in 30 μL, molecular biology grade water) and the DNA contamination was removed using the TURBO DNA-freeTM kit (Ambion® by Life TechnologiesTM) according to the manufacturer’s instructions. The RNA quantification and ratio 260/230 and 260/280 were measured by spectrophotometry (NanoDrop 2000c Thermo Scientific). The absence of DNA contamination was verified on RNA samples by PCR in RT conditions (1 ng of total RNA per reaction) on the mtrF gene (as described below).
Q(RT)-PCR
Reverse transcription reactions were carried out on 1 ng of total RNA using SuperScriptTM III Reverse Transcriptase (Life technologies) with a random primer (250 ng per reaction) according to the manufacturer’s instructions. Dilutions of 1:10 of synthesized cDNAs were used for Q-PCR. The q-PCR primers are listed in Table S1 in the supplemental material. The primers were chosen according to the bibliography and modified according to the sequence of our strain or were newly designed with the help of a primer-designing tool (Primer3 and BLAST, NCBI). For standard curve elaboration, the fragments of genes were amplified. The PCR reactions (20 μL) contained 5 ng DNA of S. algae C6G3, 1.25 μM of each primer and 10 μL of Taq’Ozyme Purple Mix 2 (Ozyme) with the following reaction parameters: 2 min at 98 °C, 30 amplifications cycles with 30 sec at 98 °C, 30 sec at 50 °C or 60 °C, 30 sec at 72 °C and ended with 5 min at 72 °C. Each q(RT)-PCR reaction (20 μL) contained 2 μL of cDNAs, 0.5 μM of each primer and 10 μL of SsoAdvancedTM SYBR® Green Supermix (Biorad) and was performed in duplicate on CFX96 Real Time System with the following reaction parameters: 30 sec at 98 °C, 35 two-step amplifications cycles with 5 sec at 98 °C and 10 sec at 62 °C or 58 °C, followed by a melting curve step to verify the specificity of the amplification product. DNA fragments were cloned into pGEMT vector (Promega) and checked by sequencing. The genes detected with RAST56 and JGI IMG/ER57,58 platforms are identified with the accession number of the JGI IMG/ER platform. After purification, the plasmid concentrations were determined by spectrophotometry and used to make the range dilution for the absolute quantification. The starting quantity (SQ) was determined using Bio-Rad CFX Manager 2.1 software and transformed into copies per milliliter of culture according to the sample dilutions. The quantification of the level of expression of the housekeeping gene (rpoD) encoding for a σ70 subunit of the RNA polymerase was used to normalize the gene expression. The normalized gene expression was averaged, and the significance of differences in expression was tested with the t-test (XLSTAT version 2010.5.02) and compared.
Enzymatic activities
Cells of S. algae C6G3 were grown anaerobically as described above until the end of the exponential phase of growth. For crude extract preparation, cells were harvested, washed three times in 10 mM TRIS-HCl buffer pH8 containing 86 mM of NaCl and 50 mM of MgSO4. Pellets were re-suspended in 50 mM TRIS buffer (pH8) and were disrupted by French press and centrifuged 15 minutes at 13 000 rpm. The crude extract corresponds to the recovered supernatant. The nitrate and nitrite reductase activities present in crude extracts were measured spectrophotometrically at 28 °C by following the oxidation of reduced benzyl viologen at 600 nm coupled to the reduction of nitrate or nitrite, respectively59. Activities were expressed as μmol of reduced NO3 or NO2/min/mg proteins. Protein concentrations were estimated by BioRad Protein assay.
Additional Information
How to cite this article: Aigle, A. et al. Physiological and transcriptional approaches reveal connection between nitrogen and manganese cycles in Shewanella algae C6G3. Sci. Rep. 7, 44725; doi: 10.1038/srep44725 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Jørgensen, B. B. Processes at the sediment–water interface., (SCOPE 21, Wiley, 1983).
Anschutz, P., Sundby, B., Lefrancois, L., Luther, G. W. & Mucci, A. Interactions between metal oxides and species of nitrogen and iodine in bioturbated marine sediments. Geochimica Et Cosmochimica Acta 64, 2751–2763, doi: 10.1016/S0016-7037(00)00400-2 (2000).
Bartlett, R., Mortimer, R. J. G. & Morris, K. M. The biogeochemistry of a manganese-rich Scottish sea loch: Implications for the study of anoxic nitrification. Cont Shelf Res 27, 1501–1509, doi: 10.1016/j.csr.2007.01.027 (2007).
Deflandre, B., Mucci, A., Gagne, J. P., Guignard, C. & Sundby, B. Early diagenetic processes in coastal marine sediments disturbed by a catastrophic sedimentation event. Geochimica Et Cosmochimica Acta 66, 2547–2558, doi: 10.1016/S0016-7037(02)00861-X (2002).
Hulth, S., Aller, R. C. & Gilbert, F. Coupled anoxic nitrification manganese reduction in marine sediments. Geochimica Et Cosmochimica Acta 63, 49–66, doi: 10.1016/S0016-7037(98)00285-3 (1999).
Luther, G. W., Sundby, B., Lewis, B. L., Brendel, P. J. & Silverberg, N. Interactions of manganese with the nitrogen cycle: Alternative pathways to dinitrogen. Geochimica Et Cosmochimica Acta 61, 4043–4052, doi: 10.1016/S0016-7037(97)00239-1 (1997).
Mortimer, R. J. G. et al. Anoxic nitrification in marine sediments. Mar Ecol-Prog Ser 276, 37–51, doi: 10.3354/Meps276037 (2004).
Mortimer, R. J. G. et al. Evidence for suboxic nitrification in recent marine sediments. Mar Ecol Prog Ser 236, 31–35, doi: 10.3354/Meps236031 (2002).
Thamdrup, B. & Dalsgaard, T. The fate of ammonium in anoxic manganese oxide-rich marine sediment. Geochimica Et Cosmochimica Acta 64, 4157–4164, doi: 10.1016/S0016-7037(00)00496-8 (2000).
Fernandes, S. O., Gonsalves, M. J., Michotey, V. D., Bonin, P. C. & LokaBharathi, P. A. Denitrification activity is closely linked to the total ambient Fe concentration in mangrove sediments of Goa, India. Estuar Coast Shelf S 131, 64–74, doi: 10.1016/j.ecss.2013.08.008 (2013).
Fernandes, S. O. et al. Anaerobic nitrification–denitrification mediated by Mn-oxides in meso-tidal sediments: Implications for N2 and N2O production. Journal of Marine Systems 144, 1–8, doi: 10.1016/j.jmarsys.2014.11.011 (2015).
Fernandes, S. O., Bharathi, P. A. L., Bonin, P. C. & Michotey, V. D. Denitrification: An Important Pathway for Nitrous Oxide Production in Tropical Mangrove Sediments (Goa, India). J Environ Qual 39, 1507–1516, doi: 10.2134/Jeq2009.0477 (2010).
Javanaud, C. et al. Anaerobic ammonium oxidation mediated by Mn-oxides: from sediment to strain level. Res Microbiol 162, 848–857, doi: 10.1016/j.resmic.2011.01.011 (2011).
Myers, C. R. & Nealson, K. H. Bacterial Manganese Reduction and Growth with Manganese Oxide as the Sole Electron-Acceptor. Science 240, 1319–1321, doi: 10.1126/science.240.4857.1319 (1988).
Kouzuma, A., Hashimoto, K. & Watanabe, K. Roles of siderophore in manganese-oxide reduction by Shewanella oneidensis MR-1. Fems Microbiol Lett 326, 91–98, doi: 10.1111/j.1574-6968.2011.02444.x (2012).
Myers, J. M. & Myers, C. R. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl Environ Microb 67, 260–269, doi: 10.1128/Aem.67.1.260-269.2001 (2001).
Myers, J. M. & Myers, C. R. Overlapping role of the outer membrane cytochromes of Shewanella oneidensis MR-1 in the reduction of manganese(IV) oxide. Lett Appl Microbiol 37, 21–25, doi: 10.1046/j.1472-765X.2003.01338.x (2003).
Aigle, A., Michotey, V. & Bonin, P. Draft-genome sequence of Shewanella algae strain C6G3. Stand Genomic Sci 10, 43 (2015).
Simpson, P. J. L., Richardson, D. J. & Codd, R. The periplasmic nitrate reductase in Shewanella: the resolution, distribution and functional implications of two NAP isoforms, NapEDABC and NapDAGHB. Microbiology 156, 302–312, doi: I 10.1099 (2010).
Chen, Y., Wang, F. P., Xu, J., Mehmood, M. A. & Xiao, X. Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. Isme J 5, 843–855, doi: 10.1038/ismej.2010.182 (2011).
Cruz-Garcia, C., Murray, A. E., Klappenbach, J. A., Stewart, V. & Tiedje, J. M. Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J Bacteriol 189, 656–662, doi: 10.1128/Jb.01194-06 (2007).
Clark, I. C., Melnyk, R. A., Engelbrektson, A. & Coates, J. D. Structure and Evolution of Chlorate Reduction Composite Transposons. Mbio 4, e00379–00313, doi: 10.1128 (2013).
Richardson, D. J. et al. The porin-cytochrome’ model for microbe-to-mineral electron transfer. Mol Microbiol 85, 201–212; doi: 10.1111/j.1365-2958.2012.08088.x (2012).
Fredrickson, J. K. et al. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6, 592–603, doi: 10.1038/Nrmicro1947 (2008).
Myers, C. R. & Myers, J. M. Cloning and sequence of cymA a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J Bacteriol 179, 1143–1152 (1997).
Marritt, S. J. et al. A functional description of CymA, an electron-transfer hub supporting anaerobic respiratory flexibility in Shewanella. Biochem J 444, 465–474, doi: 10.1042/Bj20120197 (2012).
Marritt, S. J. et al. The roles of CymA in support of the respiratory flexibility of Shewanella oneidensis MR-1. Biochem Soc T 40, 1217–1221, doi: 10.1042/Bst20120150 (2012).
Hartshorne, R. S. et al. Characterization of an electron conduit between bacteria and the extracellular environment. P Natl Acad Sci USA 106, 22169–22174, doi: 10.1073/pnas.0900086106 (2009).
Breuer, M., Rosso, K. M., Blumberger, J. & Butt, J. N. Multi-haem cytochromes in Shewanella oneidensis MR-1: structures, functions and opportunities. J R Soc Interface 12, doi: 10.1098/Rsif.2014.1117 (2015).
Marsili, E. et al. Shewanella Secretes flavins that mediate extracellular electron transfer. P Natl Acad Sci USA 105, 3968–3973; doi: 10.1073/pnas.0710525105 (2008).
Shi, L. et al. Isolation of a high-affinity functional protein complex between OmcA and MtrC: Two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J Bacteriol 188, 4705–4714, doi: 10.1128/Jb.01966-05 (2006).
Xiong, Y. J. et al. High-affinity binding and direct electron transfer to solid metals by the Shewanella oneidensis MR-1 outer membrane c-type cytochrome OmcA. J Am Chem Soc 128, 13978–13979, doi: 10.1021/Ja063526d (2006).
Shi, L., Fredrickson, J. K. & Zachara, J. M. Genomic analyses of bacterial porin-cytochrome gene clusters. Frontiers in microbiology 5, doi: 10.3389/fmicb.2014.00657 (2014).
Shi, L., Rosso, K. M., Zachara, J. M. & Fredrickson, J. K. Mtr extracellular electron-transfer pathways in Fe(III)-reducing or Fe(II)-oxidizing bacteria: a genomic perspective. Biochem Soc T 40, 1261–1267, doi: 10.1042/Bst20120098 (2012).
Meyer, T. E. et al. Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. Omics 8, 57–77; doi: 10.1089/153623104773547499 (2004).
Bartlett, R., Mortimer, R. J. G. & Morris, K. Anoxic nitrification: Evidence from Humber Estuary sediments (UK). Chem Geol 250, 29–39, doi: 10.1016/j.chemgeo.2008.02.001 (2008).
Thamdrup, B., Finster, K., Hansen, J. W. & Bak, F. Bacterial Disproportionation of Elemental Sulfur Coupled to Chemical-Reduction of Iron or Manganese. Appl Environ Microb 59, 101–108 (1993).
Beliaev, A. S. et al. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J Bacteriol 187, 7138–7145, doi: 10.1128/Jb.187.20.7138-7145.2005 (2005).
Dong, Y. Y. et al. A Crp-Dependent Two-Component System Regulates Nitrate and Nitrite Respiration in Shewanella oneidensis. Plos One 7, doi: 10.1371/journal.pone.0051643 (2012).
Smith, C. J., Nedwell, D. B., Dong, L. F. & Osborn, A. M. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microb 73, 3612–3622, doi: 10.1128/aem.02894-06 (2007).
Robertson, G. P. & Groffman, P. M. Nitrogen transformation (chapter 13). 341–364 (Springer, 2007).
Szeinbaum, N., Burns, J. L. & DiChristina, T. J. Electron transport and protein secretion pathways involved in Mn(III) reduction by Shewanella oneidensis. Env Microbiol Rep 6, 490–500, doi: 10.1111/1758-2229.12173 (2014).
Lin, H., Szeinbaum, N. H., DiChristina, T. J. & Taillefert, M. Microbial Mn(IV) reduction requires an initial one-electron reductive solubilization step. Geochimica Et Cosmochimica Acta 99, 179–192, doi: 10.1016/j.gca.2012.09.020 (2012).
Kasai, T., Kouzuma, A., Nojiri, H. & Watanabe, K. Transcriptional mechanisms for differential expression of outer membrane cytochrome genes omcA and mtrC in Shewanella oneidensis MR-1. Bmc Microbiol 15, doi: 10.1186/S12866-015-0406-8 (2015).
Bucking, C., Popp, F., Kerzenmacher, S. & Gescher, J. Involvement and specificity of Shewanella oneidensis outer membrane cytochromes in the reduction of soluble and solid-phase terminal electron acceptors. Fems Microbiol Lett 306, 144–151, doi: 10.1111/j.1574-6968.2010.01949.x (2010).
Bretschger, O. et al. Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl Environ Microb 73, 7003–7012, doi: 10.1128/Aem.01087-07 (2007).
Boogerd, F. C. & Devrind, J. P. M. Manganese Oxidation by Leptothrix-Discophora. J Bacteriol 169, 489–494 (1987).
Krumbein, W. & Altmann, H. A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgoländer Wissenschaftliche Meeresuntersuchungen 25, 347–356 (1973).
Balch, W. E., Fox, G. E., Magrum, L. J., Woese, C. R. & Wolfe, R. S. Methanogens - Re-Evaluation of a Unique Biological Group. Microbiol Rev 43, 260–296 (1979).
Baumann, P., Baumann, L. & Mandel, M. Taxonomy of marine bacteria: the genus Beneckea. J Bacteriol 107, 268–294 (1971).
Laha, S. & Luthy, R. G. Oxidation of Aniline and Other Primary Aromatic-Amines by Manganese-Dioxide. Environ Sci Technol 24, 363–373, doi: 10.1021/Es00073a012 (1990).
Misawa, T., Hashimoto, K. & Shimodaira, S. Fomation of Fe(II)-Fe(III) intermediate green complex on oxidation of ferrous ion in neutral and slightly alkaline sulphate solutions. Journal of inorganique and nuclear chemistry 35, 4167–4174 (1973).
Michotey, V. et al. Spatio-temporal diversity of free-living and particle-attached prokaryotes in the tropical lagoon of Ahe atoll (Tuamotu Archipelago) and its surrounding oceanic waters. Mar Pollut Bull 65, 525–537, doi: 10.1016/j.marpolbul.2012.01.009 (2012).
Tréguer, P. & Lecorre, P. Manuel d’analyse des sels nutritifs dans l’eau de mer. Lab. Océanogr. Chim. Univ. Bretagne Occid, Brest, 110 (1975).
Chin, C. S., Kenneth, S. J. & Kenneth, H. C. Spectrophotometric determination of dissolved manganese in natural waters with 1-(2-pyridylazo)-2-naphthol: application to analysis in situ in hydrothermal plumes. Mar Chem 37, 65–82 (1992).
Aziz, R. K. et al. The RAST server: Rapid annotations using subsystems technology. Bmc Genomics 9, doi: 10.1186/1471-2164-9-75 (2008).
Mavromatis, K. et al. The DOE-JGI Standard Operating Procedure for the Annotations of Microbial Genomes. Stand Genomic Sci 1, 63–67; doi: 10.4056/Sigs.632 (2009).
Markowitz, V. M. et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42, D560–D567, doi: 10.1093/Nar/Gkt963 (2014).
Bonin, P., Bertrand, J. C., Giordano & Gilewicz, M. Specific sodium dependence of a nitrate reductase in a marine bacterium. FEMS Microbiol Lett 48, 5–9 (1987).
Acknowledgements
This study was supported by the French National Coastal Environment Programme (PNEC “Chantier Littoral Atlantique”) and the ANR PROTIDAL project (ANR-06-BLAN-0040) and FEDER. We thank Sophie Guasco for technical help.
Author information
Authors and Affiliations
Contributions
Designing of the experiments: A.A., V.M.i., P.B., V.M.e. Performing the experiment: A.A., V.M. Enzymatic activity C.I. Redaction of the manuscript: A.A., V.M.i., P.B., V.M.e., C.I.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Aigle, A., Bonin, P., Iobbi-Nivol, C. et al. Physiological and transcriptional approaches reveal connection between nitrogen and manganese cycles in Shewanella algae C6G3. Sci Rep 7, 44725 (2017). https://doi.org/10.1038/srep44725
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44725
This article is cited by
-
Reduction of alternative electron acceptors drives biofilm formation in Shewanella algae
npj Biofilms and Microbiomes (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.