Abstract
Photoferrotrophy allows anoxygenic phototrophs to use reduced iron as an electron donor for primary productivity. Recent work shows that freshwater photoferrotrophs can use electrons from solid-phase conductive substances via phototrophic extracellular electron uptake (pEEU), and the two processes share the underlying electron uptake mechanism. However, the ability of marine phototrophs to perform photoferrotrophy and pEEU, and the contribution of these processes to primary productivity is largely unknown. To fill this knowledge gap, we isolated 15 new strains of the marine anoxygenic phototroph Rhodovulum sulfidophilum on electron donors such as acetate and thiosulfate. We observed that all of the R. sulfidophilum strains isolated can perform photoferrotrophy. We chose strain AB26 as a representative strain to study further, and find that it can also perform pEEU from poised electrodes. We show that during pEEU, AB26 transfers electrons to the photosynthetic electron transport chain. Furthermore, systems biology-guided mutant analysis shows that R. sulfidophilum AB26 uses a previously unknown diheme cytochrome c protein, which we call EeuP, for pEEU but not photoferrotrophy. Homologs of EeuP occur in a range of widely distributed marine microbes. Overall, these results suggest that photoferrotrophy and pEEU contribute to the biogeochemical cycling of iron and carbon in marine ecosystems.
Similar content being viewed by others
Introduction
Marine sediments comprise the single largest ecosystem on Earth’s surface in spatial coverage, and support nearly half of global primary productivity [1]. Geochemical studies of marine sediments have revealed that microbe-mineral electron transfer reactions are linked to carbon, sulfur, iron, and nitrogen cycling, and may play an important role in microbial energy flow [2]. Anoxygenic phototrophs are metabolically versatile and capable of utilizing both soluble and mineral forms of Fe(II) as electron donors for photosynthesis, via a process called photoferrotrophy [3]. They are known to coexist in sediments with neutrophilic iron-oxidizing (chemoautotrophic) and iron-reducing (chemoheterotrophic) bacteria, and may exchange electrons with insoluble/solid-phase substances [2] (e.g., mixed-valent iron [4,5,6] and sulfide minerals [7, 8]). Phototrophs can also use mineral proxies of solid-phase conductive substances (i.e., poised electrodes) by a process called phototrophic extracellular electron uptake (pEEU) [9, 10].
Recent bioelectrochemical studies have shown that pure cultures of freshwater photoferrotrophs, including Rhodopseudomonas palustris TIE-1, link pEEU to microbial energy generation and carbon dioxide (CO2) fixation, suggesting that primary productivity may be attributed to this process in the environment [9, 10]. Detailed biochemical and molecular work by our group has revealed electron uptake mechanisms in freshwater photoferrotrophs capable of pEEU [9, 11]. Because only a limited number of genetic markers are available to study this process, the distribution and importance of this electron uptake mechanism in marine sediments remain unclear [9, 11]. This is compounded by a lack of genetically-tractable marine photoferrotrophs in culture, which makes it challenging to understand electron uptake processes. Two marine phototrophs previously known to perform photoferrotrophy, namely Rhodovulum iodosum and Rhodovulum robiginosum are fastidious [12], and we have not been able to study them using genetic and molecular approaches thus far. However, mineralogical studies on the byproducts of photoferrotrophy produced by these microbes have been reported [6, 12]. Minerals that were produced by photoferrotrophy were identified as goethite (α-FeOOH) with minor fractions of lepidocrocite (γ-FeOOH) [6] and ferrihydrite with traces of magnetite [12]. Because anoxygenic phototrophs are prevalent in marine environments, both photoferrotrophy and pEEU may be overlooked microbial growth and survival strategies in these ecosystems.
To fill this knowledge gap, we isolated 15 related strains of Rhodovulum sulfidophilum using acetate/thiosulfate as electron donors, and further tested their ability to perform photoferrotrophy and pEEU. Remarkably, all the isolated R. sulfidophilum strains can perform photoferrotrophy. We selected R. sulfidophilum AB26 (AB26) as a representative strain to investigate further [13] using geochemical, electrochemical, systems biology, and genetic approaches. During photoferrotrophy, AB26 produces a variety of iron minerals. Bioelectrochemical studies indicate that AB26 can perform pEEU over a wide range of redox potentials (−200 mV and +100 mV) that represent the potentials of iron and iron-sulfur minerals found in marine environments. Furthermore, we show that pEEU is connected to the photosynthetic electron transport chain (pETC), and that it leads to an upregulation of genes involved in carbon dioxide (CO2) fixation. Lastly, leveraging comparative proteomics combined with mutant analysis, we identify a previously uncharacterized diheme cytochrome c protein (named EeuP here for extracellular electron uptake protein) that is important for pEEU but not photoferrotrophy. EeuP homologs can be found across bacteria including organisms belonging to the ecologically important marine Roseobacter group. These findings suggest that both photoferrotrophy and pEEU might be more prevalent in marine ecosystems than previously thought. Our results offer new opportunities to understand these fundamental biological processes that shape our planet.
Materials and methods
Microbial isolation and cultivation conditions
Rhodovulum sulfidophilum strains (including AB26 (AB26)) were isolated in July 2014 from independent microbial mat samples from the Trunk River estuary in Woods Hole, MA. Enrichments were cultivated photoheterotrophically in anoxic artificial seawater (SW) medium supplemented with 20 mM acetate or 10 mM sodium thiosulfate. Enrichments were cultivated with ∼850-nm light at 30 °C and passaged six times. To isolate colonies, cultures from the anaerobic enrichments were streaked aerobically on Bacto agar with Difco marine broth (MB) 2216 (BD Diagnostic Systems, Cockeysville, MD, USA). The individual colonies were passaged six more times by streaking on MB-agar plates and used for further characterization. Further details can be found in the supplemental materials.
Cell suspension assay
The cell suspension assay was performed in an anaerobic chamber (80% N2, 15% CO2, and 5% H2, Coy Laboratory, Grass Lake, MI, USA) at room temperature using photoautotrophically grown R. sulfidophilum strains with hydrogen as previously described [14]. Further details can be found in the supplemental materials.
Bulk BES setup and conditions
Bioelectrochemical systems (BESs) were configured as previously described [10, 15]. Further details can be found in the supplemental materials.
Next-generation microfluidic bioelectrochemical cell (µ-BEC) and conditions
µ-BEC arrays were miniaturized BESs consisting of 2 × 6 fluidically connected microchambers (~1 µL each) that enabled electrochemical measurements in continuous flow or batch mode of operation. Further details can be found in the supplemental materials.
Construction of ∆eeuP (BV509_10070) AB26 mutant strain
To develop a eeuP deletion mutant of AB26, pJP5603-DG1, a suicide vector, was constructed from pJP5603 [16, 17]. Further details can be found in the supplemental materials.
Results
Multiple isolates of the marine bacterium Rhodovulum sulfidophilum can perform photoferrotrophy
To expand our understanding of microbe-mineral electron transfer reactions, we isolated 15 phototrophic bacteria from an estuarine microbial mat (Trunk River, Woods Hole, MA, USA) using acetate/thiosulfate as electron donors and sequenced their genomes (Supplementary Fig. 1). The Trunk River isolates were ~99% related to the type-strain of R. sulfidophilum (strain DSM 2351), a marine phototroph, based on analysis of their photosynthetic reaction center M (pufM) and 16 S rDNA gene sequences. Fifteen R. sulfidophilum strains were isolated from microbial mat enrichments from independent microbial mat samples from Trunk River, and were capable of typical purple nonsulfur bacterial growth modes, including anaerobic (photoheterotrophy and photoautotrophy) and aerobic (chemoheterotrophic) growth (Fig. 1a, Supplementary Table 1). To screen the isolates for their ability to oxidize Fe(II), we cultivated them under illuminated conditions with Fe(II)-NTA (nitrilotriacetic acid) as the sole electron donor. We observed complete oxidation of Fe(II)-NTA in these incubations for all strains (Fig. 1b; Supplementary Table 1). We chose to investigate R. sulfidophilum AB26 (AB26) further based on its ease of cultivation on liquid and solid media, which makes it favorable for downstream genetic and synthetic biological manipulation.
a Aerobic chemoheterotrophic growth, anaerobic photoheterotrophic growth (acetate), and photoautotrophic growth (thiosulfate or H2 as the electron donor). Error bars are means ± standard deviations of three biological replicates. b Fe(II)-NTA oxidation by AB26 in cell suspension assays. Error bars represent means ± standard deviations of three biological replicates assayed in duplicate.
To investigate whether Fe(II) oxidation contributes to the growth of AB26, we performed physiological growth experiments using Fe(II)-NTA. AB26 completely oxidized 5 mM Fe(II)-NTA within 3.5 days with a corresponding increase in optical cell density (OD660) and total protein content (Fig. 2a, b and Supplementary Fig. 2), indicating that phototrophic Fe(II)-oxidation is connected to cellular growth. Scanning electron microscopy (SEM) coupled with energy-dispersive X-ray spectroscopy (EDS) of samples from these incubations revealed the formation of iron-phosphate minerals closely associated with AB26 cells (Fig. 2c). AB26 also oxidized Fe(II) in the absence of NTA, but with a ~20-fold lower rate (~80 µM day−1) (Fig. 2a and d). We observed an increase in total protein content in these incubations (Fig. 2e), indicating that AB26 can use non-chelated Fe(II) as its sole electron donor for photoautotrophic growth. X-ray diffraction (XRD) showed the formation of divalent, mixed valent, and trivalent iron minerals such as siderite (FeCO3), vivianite [(Fe3(PO4)2•8(H2O)], green rust carbonate [GR(CO32−)], goethite (FeOOH), and traces of akaganeite (FeOOH, Cl) (Fig. 2f). Together, these results highlight the ability of the marine bacterium R. sulfidophilum AB26 to perform Fe(II)-oxidation and connect this process to photoautotrophic growth.
a Fe(II)-NTA oxidation by AB26 and b corresponding growth determined as OD660 during photoferrotrophy. Error bars are means ± standard deviations of three biological replicates. No cell and heat-killed controls are shown. c Scanning electron micrograph (SEM) and energy-dispersive X-ray spectroscopy (EDS) of cell-associated iron phosphate produced during Fe(II)-NTA oxidation by AB26. d Fe(II) oxidation without NTA by AB26 and e corresponding total protein content. Error bars represent means ± standard deviations of three biological replicates. f X-ray diffraction patterns of the oxidation products obtained from Fe(II)no-NTA oxidation. XRD indicated the formation of divalent, mixed valent, and trivalent iron minerals such as siderite (S), vivianite (V), goethite (G), green rust (GR), and akageneite (A) after the end of incubation: (black: no cell control; blue: with AB26). Inset: SEM image showing the iron minerals present in the reactor.
AB26 accepts electrons from a poised electrode
A fundamental knowledge gap is whether marine photoferrotrophs can also access electrons from insoluble or solid-phase conductive substances for cellular metabolism. To investigate this question, we utilized bioelectrochemical systems (BESs). BESs mimic microbial interactions with insoluble minerals, wherein an electrode can operate as an electron donor or acceptor for microbial metabolism [9, 10].
We cultivated AB26 photoautotrophically in artificial seawater medium with indium tin oxide (ITO) as working electrodes. Because photoferrotrophs have been previously observed to perform pEEU at the midpoint potential of insoluble iron minerals (~+100 mV) [18], we poised the electrodes at +100 mV vs. Standard Hydrogen Electrode (SHE). We observed current densities of ~−13.4 ± 0.5 nA cm−2 under phototrophic conditions compared to abiotic controls (Fig. 3a, Supplementary Table 2). In marine ecosystems, high sulfate concentrations (~28 mM) favor the formation of iron-sulfide minerals [19, 20] that generally have lower redox potentials than soluble iron [21]. To assess whether AB26 can accept electrons at lower redox potentials, we poised electrodes at −200 mV vs. SHE. At −200 mV, we observed current densities of ~−16.8 ± 0.6 nA cm−2 (Fig. 3a, Supplementary Table 2). These current densities are ~25% higher when compared to current densities observed at +100 mV, suggesting that −200 mV may be a preferable potential for pEEU.
a Electron uptake (represented as current density) by wild-type AB26 compared to abiotic controls in the BES under illuminated conditions. The data shown are representative of three experiments. b Cyclic voltammograms (CV) after 108 h of electrochemical incubations compared to an abiotic control at −200 mV versus SHE. c LIVE/DEAD staining of the ITO-electrode after electrochemical incubation in BES at −200 mV versus SHE. Green indicates live cells. d Electron uptake by wild-type AB26 at −200 mV versus SHE in the µ-BEC under light-dark cycle (shaded region represents dark condition) compared to abiotic controls. The data shown are representative of three experiments. For the source data, refer to Supplementary Table 2.
Cyclic voltammetry (CV) of the electrodes at both +100 mV and −200 mV vs. SHE revealed two distinct cathodic peaks at −50 and −430 mV (Fig. 3b, Supplementary Fig. 3). These results suggest the presence of redox-active components in the system indicative of microbial redox activity. Consistent with current density measurements, CV peaks are also higher for electrodes poised at −200 mV vs. SHE. Confocal fluorescence microscopy showed the presence of viable AB26 cells on electrodes, suggesting AB26 can utilize poised electrodes for cellular survival (Fig. 3c, Supplementary Fig. 3). Together, these results suggest that AB26 can take up electrons from electrodes poised over a range of potentials that mimic the redox potentials of naturally abundant iron and sulfur minerals.
pEEU is connected to the photosynthetic electron transport
To investigate whether pEEU is linked to the photosynthetic electron transfer in AB26, we performed physiological experiments using well-characterized inhibitors in microfluidic BESs that were scaled for sensitive electrochemical measurements, i.e., microfluidic bioelectrochemical cells (µ-BECs). These systems have the advantage of a higher signal-to-noise ratio than traditional bulk BESs and allow the response of surface-attached cells to be isolated because planktonic cells can be washed out with microfluidic control [10].
We advanced an earlier µ-BEC prototype to create arrays that can be scaled to 96-wells. The new platform is superior to that reported previously regarding ease of assembly and use, suitability for in situ microscopy, and throughput [10]. Surface micromachining was used to fabricate µ-BEC electrodes, and fluidic channels were etched in glass allowing for smaller reaction chambers and higher reproducibility in the present study. The µ-BECs were used to study pEEU by AB26 (Supplementary Fig. 4) (see Methods section for a complete description of the µ-BEC design, microfabrication, and assembly). Once current densities (~−30 nA cm−2) were stabilized under constant illumination, the planktonic cells were washed out of the system with microfluidic control. From surface-attached cells, we observed similar current densities at −200 mV (~−30.2 ± 0.1 nA cm−2; Fig. 3d), and +100 mV (~−31.2 ± 0.7 nA cm−2; Supplementary Fig. 3c), respectively. This observation was in contrast to the bulk-BESs, where we observed increased current densities at −200 mV (Fig. 3a). This difference might be due to the presence of planktonic cells in bulk BESs and/or differential cell attachment in the two systems. The current densities for AB26 were lower when compared to those we observed in R. palustris TIE-1 using the previous format of the µ-BEC [9, 10].
To test whether the electron flow during pEEU is linked to photosynthesis in AB26, we selectively inhibited key-steps in cyclic pETC (Fig. 4a, Supplementary Fig. 5a) using a chemical probe-based approach [10]. To determine whether the photosynthetic reaction center (RC) is essential for pEEU, we first exposed AB26 to terbutryn (2-tert-butylamino-4-ethylamino6-methylthio-1,3,5-triazine), an inhibitor that blocks electron transfer from QA to QB in RCs of purple bacteria, cyanobacteria, and higher plants [22,23,24,25,26,27,28]. We observed an ~80% decrease in current density under illuminated conditions upon terbutryn treatment at both −200 mV (−7.3 ± 0.08 nA cm−2) and +100 mV (−6.2 ± 0.9 nA cm−2) (Fig. 4b, Supplementary Fig. 5b, Supplementary Tables 3 and 4). There were no differences in current densities between treated and untreated controls under dark conditions (Fig. 4b). These results demonstrate that the RC of AB26 is critical for pEEU.
a Schematic diagram of the site of inhibition (indicated by a red halo) in the electron path of the photosynthetic ETC (pETC). P870 (photosystem), P870* (excited photosystem), UQ (ubiquinone), bc1 (cytochrome bc1), c2 (cytochrome c2), NADH-DH (NADH dehydrogenase), Δp (proton gradient), H+ (protons), hν (light),? (currently unknown), PMF (proton motive force) and ATP (adenosine triphosphate). Letters b-e in parenthesis indicate inhibition sites of different inhibitors used. Electron uptake (represented as current density) by WT AB26 in the μ-BEC under a light-dark cycle (shaded region) with (red) and without (blue) exposure to pETC inhibitors, b terbutryn, c carbonyl cyanide m-chlorophenyl CCCP), d rotenone, and e antimycin A, respectively. The electrode was poised at −200 mV versus SHE in these experiments. Data shown are representative of three experiments. For the source data, refer to Supplementary Table 3.
Cyclic electron flow by the pETC generates a proton motive force (PMF) that is necessary to drive ATP production in anoxygenic phototrophs [29]. To investigate whether a proton gradient is important for pEEU, we treated biofilms with carbonyl cyanide m-chlorophenyl hydrazine (CCCP). CCCP is a protonophore that dissipates the proton gradient and uncouples it from ATP synthesis [10, 29, 30]. We observed ~80% and ~90% decreases in current densities under illuminated conditions upon CCCP treatment at −200 mV (−5.9 ± 0.1 nA cm−2) and +100 mV (−3.3 ± 0.1 nA cm−2), respectively (Fig. 4c, Supplementary Fig. 5c, Supplementary Tables 3 and 4). Similar to terbutryn treatment, CCCP treatment did not affect current density under dark conditions (Fig. 4c). These results demonstrate that establishing a PMF across the inner membrane is important for pEEU.
The energy-transducing (proton-translocating) NADH dehydrogenase (complex I) generates a PMF for ATP production by oxidizing NADH. NADH dehydrogenase can also dissipate the PMF to couple electrons from the ubiquinone pool to NAD+ reduction using a process called reverse electron flow [10, 31, 32]. To determine whether NADH dehydrogenase is important for pEEU, we treated biofilms with the NADH dehydrogenase inhibitor rotenone [33]. Rotenone works by blocking electron transfer from the iron-sulfur clusters in NADH dehydrogenase to ubiquinone [34]. We observed a ~70% decrease in current uptake in rotenone-treated biofilms under illuminated conditions at both −200 mV (−9.9 ± 0.05 nA cm−2) and +100 mV (−9.2 ± 0.07 nA cm−2) (Fig. 4d, Supplementary Fig. 5d, Supplementary Tables 3 and 4). These results suggest that electrons are cycled to NADH dehydrogenase and may be important for the generation of cellular reducing equivalents.
The proton-translocating cytochrome bc1 is a key component of the pETC. To investigate whether cytochrome bc1 has a role in pEEU, we treated AB26 with antimycin A which inhibits electron transfer from ubiquinol to cytochrome b [35, 36]. We observed a ~25% decrease in current uptake upon antimycin A treatment in treated biofilms compared to untreated controls at both −200 mV (−22.2 ± 0.1 nA cm−2) and +100 mV (−23.3 ± 1.3 nA cm−2) (Fig. 4e, Supplementary Fig. 5e, Supplementary Tables 3 and 4). Notably, the magnitude of the decrease in current uptake increased in each consecutive light-dark cycle (Fig. 4e). DMSO (dimethyl sulfoxide) was used as the solvent for all the chemical inhibitors used in these physiological experiments, and we did not observe any effect on current uptake by DMSO treatment alone (Supplementary Fig. 6 and Supplementary Table 5). These results indicate that cytochrome bc1 is important for pEEU.
Transcriptomic analysis of potential electron sinks during pEEU
To assess whether carbon metabolism is activated during pEEU, and to identify the potential electron sinks in AB26, we performed genome-wide transcriptome analysis compared to aerobic growth. We observed an upregulation of form I ruBisCO (~2-fold; P < 0.05), but not form II ruBisCO during pEEU (Fig. 5a, Supplementary Table 6). We also observed the upregulation of the putative LysR-family transcriptional regulator, CbbR [37] (BV509_05530 and BV509_15210). We observed that the cbbR homolog adjacent to form I ruBisCO is highly expressed during pEEU, even when compared to other phototrophic growth conditions (Fig. 5a). CbbR is known to activate the transcription of form I ruBisCO in response to the redox, energy, and carbon status of the cell [38,39,40]. This data suggests that AB26 may utilize the Calvin–Benson–Bassham (CBB) cycle (that uses RuBisCO) as an electron sink for pEEU and that CbbR may play a role in regulating CBB cycle activity.
a Expression analysis of genes encoding the Calvin-Benson-Bassham cycle and b potential carbon storage pathways in the genome compared to aerobic growth. RuBP (Ribulose 1,5-bisphosphate), 1,3 BPG (1,3-bisphosphoglycerate), G3P (Glyceraldehyde 3-phosphate), FBP (Fructose 1,6-bisphosphate), F6P (Fructose 6-phosphate), X5P (Xylulose 5-phosphate), Ru5P (Ribulose 5-phosphate) and R5P (Ribose 5-phosphate). Source data (and reactions not mentioned in the text) are provided as a Source Data File.
We also observed the upregulation of a gene cluster that encodes a putative formate dehydrogenase (BV509_15145-BV509_15165) under pEEU compared to photoautotrophically grown cells with hydrogen (H2) (i.e., the inoculum for pEEU experiments). Formate dehydrogenase catalyzes formate oxidation to CO2 and H+ but can also function in reverse to catalyze CO2 fixation to produce formate [41, 42]. CO2 fixation via formate dehydrogenase has been observed in Rhodobacter capsulatus [41] and other microbes [43]. Studies have also shown that formate dehydrogenase is an electrochemically active enzyme that can participate in direct electron uptake from electrodes [44, 45]. The AB26 formate dehydrogenase cluster shows high similarity to the fdsGBACD operon that encodes the cytoplasmic molybdenum-containing formate dehydrogenases in Rhodobacter species (Supplementary Fig. 7). All five genes of this cluster were highly upregulated (~4-fold; P < 0.0001) during pEEU (Supplementary Table 7).
Because of their role in the microbial carbon cycle, we identified putative carbon storage pathways and assessed their expression. The polyhydroxybutyrate (PHB) biosynthesis enzymes, including a PhaC homolog responsible for PHB polymerization, were expressed but typically downregulated during pEEU (Fig. 5b). A polyhydroxyalkanoate (PHA) synthesis repressor phaR homolog (BV509_06285) and a PHA depolymerase phaZ homolog (BV509_06270) were upregulated during pEEU (Fig. 5b). In addition, a phasin-like protein was highly upregulated during pEEU (~4-fold; P < 0.0001) (Fig. 5b). Phasins localize to the surface of PHB granules and are synthesized under conditions favorable for PHB production [46]. Lastly, a putative glycogen biosynthetic gene cluster was identified. The glycogen biosynthesis pathway was expressed at low levels under all phototrophic growth conditions (Fig. 5b).
We screened the transcriptome for additional electron-consuming pathways, in particular, putative terminal oxidases that could be important in the oxygen-limiting conditions phototrophs are exposed to in marine sediments [47, 48]. We observed the upregulation of a gene cluster (BV509_18680-BV18695) encoding a cbb3-type oxidase (2–3.5-fold; P < 0.0001) under pEEU compared to H2-grown cells. The activity of these enzymes might explain the partial decrease in pEEU upon cytochrome bc1 inhibition. We also observed upregulation of cytochrome c peroxidases (BV509_14915, ~6-fold; P < 0.0001) (Supplementary Table 7), a common cellular stress response in bacteria [49]. Together, these results show that alternative ubiquinone oxidases are upregulated during pEEU, which may provide additional electron sinks in oxygen-limited environments.
AB26 uses a diheme cytochrome c protein, EeuP, for pEEU but not photoferrotrophy
Microbes exchange electrons with insoluble/solid-phase electron donors and acceptors via both direct and indirect means. EET pathways typically involve c-type cytochromes [50] or iron–sulfur cluster containing electron transfer proteins to mediate iron oxidation [51, 52] and/or EEU [53]. To better understand the underlying EEU mechanisms in AB26, we used systems biology approaches to identify potential electron transfer proteins involved in pEEU for downstream biochemical and molecular genetic analysis.
We analyzed the annotated genome of AB26 for the presence of potential electron transfer proteins, specifically cytochrome c and iron-sulfur proteins, and assessed their expression. Cytochrome c proteins ranging from monoheme (Cyc2), diheme (FoxE), and decaheme (PioA/MtoA) have been implicated in pEEU [50, 54]. The AB26 genome encodes several putative cytochrome c and iron-sulfur proteins based on the identification of canonical protein motifs (Supplementary Tables 8 and 9). However, its genome does not encode homologs of electron transfer proteins that have been characterized in known iron oxidation/reduction [55] and/or extracellular electron transfer (EET) pathways [50, 54]. We identified a total of 40 putative cytochrome c-like proteins utilizing the signature CXXCH motif (heme-binding site) as a query [56] (Supplementary Table 8). Among these proteins, only 22 are predicted to contain Sec signal peptides or transmembrane helices by PredTat [57]. Many of the cytochrome c-like proteins identified are homologs of well-characterized c-type cytochromes, such as those involved in sulfur oxidation (soxA, soxX) [58], cellular detoxification (cytochrome c peroxidase family) [59], or respiratory electron transfer (cytochrome cbb3-type and cytochrome bd). We also identified 13 genes encoding putative iron-sulfur proteins in the genome of AB26 (Supplementary Table 9).
We next assessed the genome-wide expression of the putative electron transfer proteins identified using differential expression analysis. We observed an upregulation of at least five putative c-type cytochromes with a log2-fold increase of >2 under pEEU compared to photoautotrophic growth with H2. This included subunits of a putative cbb3-type oxidase (BV509_18680, ~2-fold; P < 0.0001 and BV509_18690, ~3-fold; P < 0.0001), one cytochrome c peroxidase (BV509_14915, ~6-fold; P < 0.0001), and two monoheme cytochrome c proteins (BV509_18570, ~2-fold; P < 0.0001 and BV509_04560, ~2-fold; P < 0.0001). In additional, several diheme c-type cytochromes near gene clusters encoding metal transport function were upregulated under pEEU compared to H2-growth. This includes BV509_10070, which is a diheme cytochrome c near gene clusters encoding molybdenum, nickel, and cobalt transport systems (~2-fold; P < 0.0001).
One common feature of EET pathways in Gram-negative bacteria is that they require c-type cytochromes to facilitate electron transfer across the outer membrane [50, 54]. Subsequently, we investigated the protein expression of cytochrome c proteins in AB26 during photoferrotrophy and pEEU using heme-staining followed by protein mass spectrometry. We observed at least three heme-stainable protein bands that were enriched during both photoferrotrophy and pEEU, compared to H2-grown cells (Fig. 6). Corresponding heme-stainable bands were excised from the Coomassie-stained gel. Proteins were in-gel digested with trypsin and analyzed by mass spectrometry. Mass spectrometry analysis of these heme-stainable bands revealed multiple putative cytochrome c proteins (Table 1). Based on the abundance and the molecular size of these heme stainable protein bands, we identified two top hits: a putative diheme (BV509_10070) and a triheme (BV509_00325) cytochrome c. BV509_00325 has 94% identity to a well-characterized photosynthetic reaction center cytochrome c subunit PufC [60], whereas the diheme cytochrome c-like gene BV509_10070 has no characterized homolog.
Coomassie and Heme staining of total soluble and membrane proteins of AB26 harvested under Fe(II) oxidation (a) and pEEU (b). Asterisks represent upregulated heme protein bands (black, 1; blue, 2; red, 3) in either Fe(II)-exposed or pEEU conditions. These enriched heme bands were analyzed by mass spectrometry (Table 1). Hydrogen grown cells were used as inoculum and as a negative control. Near equal protein-loadings were confirmed by Coomassie stain.
We chose to characterize BV509_10070 at the genetic level due to its unknown function and increased protein expression during photoferrotrophy and pEEU. BV509_10070, hereafter named EeuP is a 573 amino acid diheme cytochrome c with a putative Sec signal peptide (Fig. 7a). To better understand the role of EeuP, we developed a genetic system de novo in AB26 and generated an eeuP deletion mutant (ΔeeuP). The ΔeeuP mutant had no growth defects under either aerobic (chemoheterotrophic), or anaerobic conditions such as photoheterotrophy on acetate and photoautotrophy using H2/thiosulfate compared to the wild-type (WT) (Supplementary Fig. 8, Supplementary Table 10). The mutant was also confirmed to not contain EeuP by analyzing total protein using heme staining and mass spectrometry analysis (Supplementary Fig. 9, Supplementary Table 11). We also did not detect a defect in phototrophic Fe(II)-NTA oxidation either in cell suspension assays or during physiological growth experiments, when compared to the WT (Supplementary Fig. 10a–c).
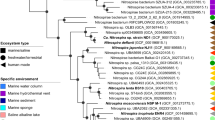
a Working schematic representation of EeuP protein sequence inlcuding putative functional domains: Sec signal peptide (purple), cytochrome c domain (red), heme-binding motif (orange). b The site of inhibition in the electron path of pETC is indicated by a red halo. P870 (photosystem), P870* (excited photosystem), UQ (ubiquinone), bc1 (cytochrome bc1), c2 (cytochrome c2), NADH-DH (NADH dehydrogenase), Δp (proton gradient), H+ (protons), hν (light),? (currently unknown), PMF (proton motive force) and ATP (adenosine triphosphate). c Electron uptake (represented as current density) by WT AB26 and the ΔeeuP mutant in the μ-BEC under a light-dark cycle. The electrode was poised at −200 mV versus SHE in these experiments. Data shown are representative of three experiments. For the source data, refer to Supplementary Table 12. d Phylogenetic supergene tree consisting of concatenated recA-rpoB-dnaK protein sequences of 206 bacteria and two archaea sequences as the root (orange), consisting of four bacterial phyla. Bold lines on the tree signify bootstrapping values > = 90. Species containing BLASTP-identified homologous protein sequences of eeuP are identified via yellow-filled circles at the ends of nodes. Homologs of eeuP are identified in the Alpha-, Beta-, and Gammaproteobacteria orders, and one homolog within the Acidobacteria.
We next tested whether EeuP is important for pEEU in µ-BECs (Fig. 7b, c). Interestingly, we observed a ~76% decrease in current uptake for the ΔeeuP mutant (−7.29 ± 1.87 nA cm−2) under illuminated conditions at −200 mV vs. SHE, when compared to the WT (−30.66 ± 0.82 nA cm−2) (Fig. 7c, Supplementary Table 12). A similar decrease in current density was observed in bulk BESs (~68%; Supplementary Fig. 11 and Supplementary Table 13). The defect in current uptake in the ΔeeuP mutant was restored by complementation of eeuP in trans from a plasmid (Supplementary Fig. 12 and Supplementary Table 14). These results indicate that the diheme cytochrome c, EeuP, is important for pEEU but that its activity is not required for Fe(II) oxidation.
To determine the taxonomic distribution of EeuP, we performed BLASTP analysis, and probed potential homologs for the presence of diheme motifs (Fig. 7a). Diheme-containing amino acid sequences identified were overlayed onto a concatenated supergene tree, represented as yellow-filled circles at the end of nodes (Fig. 7d, Supplementary Fig. 13). Our phylogenetic analysis of EeuP identified 56 homologs present in two phyla: Proteobacteria and Acidobacteria (Supplementary Fig. 13). Only a single homologous sequence was detected within the Acidobacteria in the order Blastocatellales. Acidobacteria, although underrepresented in culture, are metabolically diverse and ubiquitous in the environment [61]. The majority of EeuP homologs were in the Proteobacteria. There are multiple orders represented in the Alphaproteobacteria, including other members of the marine genus Rhodovulum [62, 63] (to which AB26 belongs [13]), and various members of the marine Leisingera species that belong to the Roseobacter group (which account for >20% of marine phytoplankton [64]). Homologs of EeuP were also present in soil methane-oxidizing and nitrogen-fixing Rhizobiales. The Betaproteobacterial species containing EeuP homologs include the chemolithoautotrophic Nitrosomonadaceae family [65]. Gammaproteobacterial species that possess EeuP homologs include marine species such as Alteromonas ponticola, and Nitrosococcus oceani, a gammaproteobacterial ammonia-oxidizing bacterium (AOB). In additional, EeuP homologs were identified in strains of the human pathogen Pseudomonas aeruginosa.
This analysis indicates that EeuP is widely distributed among marine bacteria. Further studies are required to evaluate the role of EeuP in these microbes. In addition, further work is necessary to assess the role of the other cytochrome c proteins identified in the mass spectrometry analysis (Table 1). These analyses may provide insights into the mechanism of pEEU in AB26 and other marine bacteria.
Discussion
Microbial iron oxidation is connected to carbon, sulfur, and nitrogen biogeochemical cycling in marine sediment environments [2]. These processes are catalyzed by neutrophilic Fe(II)-oxidizing chemoautotrophic and photoautotrophic bacteria. Recent studies have characterized marine chemoautotrophic Fe(II)-oxidizers using molecular genetic, physiological, and bioelectrochemical approaches [54]. However, relatively little is known about the ecological role of photoautotrophic Fe(II)-oxidizers or photoferrotrophs in marine ecosystems, and whether these organisms can also use solid-phase electron donors for cellular metabolism.
Redox-active solid-phase substances such as iron and iron-sulfur minerals are ubiquitous in natural environments. Microbes that exchange electrons with these minerals play an important role in the biogeochemical cycling of elements in nature. Recent studies have shown that anoxygenic phototrophs can use insoluble electron donors for CO2 fixation via phototrophic extracellular electron uptake (pEEU). However, the ecological role of photoferrotrophy and pEEU in marine ecosystems, and their possible contribution to the biogeochemical cycling of iron and carbon is poorly understood. To better understand the role of photoferrotrophy and pEEU in natural environments, we isolated 15 strains of marine purple nonsulfur bacteria, Rhodovulum sulfidophilum. Using the representative strain AB26, here we show that R. sulfidophilum is capable of both photoferrotrophy and pEEU. AB26 uses a previously uncharacterized diheme cytochrome c protein, EeuP, for pEEU but not photoferrotrophy.
Our data show that AB26 is capable of photosynthetic growth using both Fe(II) (NTA chelated or non-chelated) (Fig. 2) and poised electrodes (Fig. 3). Studies suggest that Gram-negative phototrophs oxidize soluble iron extracellularly because the product of this process is insoluble, which is lethal if accumulated in the periplasm [66,67,68,69]. We observed that oxidation of chelated or non-chelated Fe(II) by AB26 produced morphologically and chemically distinct minerals (Fig. 2c, f). Oxidation of chelated Fe(II) produced Fe(III)-phosphate minerals closely associated with AB26 cells (Fig. 2c). The presence of phosphate (1 mM) in the seawater medium may have favored the formation of poorly-crystalline Fe(III)-phosphate minerals. In contrast, oxidation of non-chelated Fe(II) led to the precipitation of divalent [Fe(II)], mixed valent [Fe(II)-Fe(III)], and trivalent [Fe(III)] iron minerals including siderite, vivianite, goethite, and green rust (Fig. 2f). The formation of Fe(II) minerals such as siderite and vivianite could be the result of the initial abiotic reaction of ferrous iron with bicarbonate or phosphate in the medium, respectively. Green rust has been shown to form as an intermediate phase during the oxidation of Fe(II) to Fe(III) minerals by Acidovorax sp. BoFeN1 [70], Klebsiella mobilis [71], and Culture KS [72]. The formation of this metastable mineral likely indicates a biomineralization pathway via precipitation of green rust by AB26 cells followed by solid-state transformation into Fe(III) minerals such as goethite.
A previous Fe(II) oxidation study using the marine phototroph R. iodosum also shows the formation of amorphous mineral phases followed by the precipitation of more crystalline goethite minerals [6]. Interestingly, despite a close association of AB26 cells with mineral aggregates, we found that the cells do not become encrusted by the minerals (Fig. 2c, f). It is possible that exopolymeric substances (EPS) may play a critical role in preventing cell encrustation, likely by binding Fe(III), similar to the activity observed by R. iodosum [6]. Although we identified pathways encoding EPS biosynthesis in the AB26 genome, further studies will be required to confirm the role of EPS in biomineralization. These observations highlight the prevalence and significance of this metabolism in marine sediments. For example, direct enzymatic Fe(III) reduction by iron-reducing bacteria has been shown to be substantial, accounting for as much as 90% of the oxidation of organic matter in marine sediments [73]. Thus, the microbially-produced Fe(III) minerals could serve as electron sinks for both chemical and biological processes, contributing to the iron cycle in marine environments.
Inhibitor studies (Fig. 4 and Supplementary Fig. 5) indicate that the pETC is important for pEEU, and that electrons are likely utilized to reduce NAD+ via NADH dehydrogenase. Interestingly, we observed only a partial decrease in pEEU upon inhibition of the cytochrome bc1 complex. This result could be due to the following reason. Cytochrome bc1 in AB26 may have a natural resistance to antimycin A, which has been described in anoxygenic phototrophs, such as Rubrivivax gelatinosus [74]. Amino acid substitutions generally confer antimycin resistance in the cytochrome b subunit of cytochrome bc1 (PetB) that displaces quinone from the Qi site. Site-directed mutagenesis of the Qi sites of PetB has been previously shown to contribute to antimycin resistance [75, 76]. Specifically, substitutions in the amino acids I49F, A55M, and I254T in the Qi pocket of PetB in R. gelatinosus compared to R. sphaeroides have been implicated in antimycin resistance [74]. We identified that the AB26 PetB homolog (BV509_00830) contains at least two amino acid substitutions (A55F and I254V) [74] (Supplementary Fig. 14). These amino acid substitutions could be responsible for the partial antimycin A-resistance we observe in AB26.
Similar to the genomes of other marine photoferrotrophs, such as R. robiginosum [11], the AB26 genome does not contain homologs of previously identified proteins that are involved in either iron oxidation/reduction [55] and/or EET [50, 54]. This result suggests that AB26 may utilize a distinct and new EEU pathway. We identified several cytochrome c proteins that were enriched specifically during photoferrotrophy and pEEU (Table 1). We tested the role of one such protein, a diheme cytochrome c-like protein (EeuP), by developing an eeuP gene deletion mutant. We were able to successfully develop a genetic system for this newly isolated marine photoferrotroph and show that EeuP is important for pEEU in AB26 (Fig. 7c, Supplementary Figs. 8–12). The ΔeeuP mutant showed a significant loss of its ability to perform pEEU but did not show any defect in photoferrotrophy. This result might suggest that, unlike freshwater photoferrotrophs, AB26 contains distinct mechanisms for these two metabolisms. It is plausible that there is functional redundancy in iron oxidase or electron transfer proteins used by AB26. Consistent with this hypothesis, we observed upregulation of additional cytochrome c proteins in the ΔeeuP mutant (Supplementary Fig. 9 and Supplementary Table 11).
Using whole-genome transcriptome analysis, we provide new insights into the molecular and bioenergetic pathways that enable photoautotrophs to utilize insoluble/solid-phase electron donors for cellular survival. This work also expands the known diversity of organisms capable of photoferrotrophy and pEEU, and provides a genetically-tractable bacterium for use in future electrochemical and physiological studies. Because AB26 utilizes electrodes poised at sufficiently more negative potentials than purple nonsulfur bacteria previously characterized, this microbe may have utility in applications including energy storage, carbon capture, and microbial electrosynthesis [77]. Future studies will characterize the photoferrotrophy and pEEU pathways, and examine how this process is connected to photosynthesis and CO2 fixation. In sum, this study opens new doors for studies on EEU in marine microbes more broadly.
References
Snelgrove PVR, Thrush SF, Wall DH, Norkko A. Real world biodiversity–ecosystem functioning: a seafloor perspective. Trends Ecol Evol. 2014;29:398–405.
Otte JM, Harter J, Laufer K, Blackwell N, Straub D, Kappler A, et al. The distribution of active iron-cycling bacteria in marine and freshwater sediments is decoupled from geochemical gradients. Environ Microbiol. 2018;20:2483–99.
Widdel F, Schnell S, Heising S, Ehrenreich A, Assmus B, Schink B. Ferrous iron oxidation by anoxygenic phototrophic bacteria. Nature. 1993;362:834–6.
Byrne JM, Klueglein N, Pearce C, Rosso KM, Appel E, Kappler A. Redox cycling of Fe(II) and Fe(III) in magnetite by Fe-metabolizing bacteria. Science. 2015;347:1473–6.
Byrne JM, van der Laan G, Figueroa AI, Qafoku O, Wang C, Pearce CI, et al. Size dependent microbial oxidation and reduction of magnetite nano- and micro-particles. Sci Rep. 2016;6:30969.
Wu W, Swanner ED, Hao L, Zeitvogel F, Obst M, Pan Y, et al. Characterization of the physiology and cell-mineral interactions of the marine anoxygenic phototrophic Fe(II) oxidizer Rhodovulum iodosum–implications for Precambrian Fe(II) oxidation. FEMS Microbiol Ecol. 2014;88:503–15.
Kondo K, Okamoto A, Hashimoto K, Nakamura R. Sulfur-mediated electron shuttling sustains microbial long-distance extracellular electron transfer with the aid of metallic iron sulfides. Langmuir. 2015;31:7427–34.
Hernandez ME, Newman DK. Extracellular electron transfer. Cell Mol Life Sci. 2001;58:1562–71.
Gupta D, Sutherland MC, Rengasamy K, Meacham JM, Kranz RG, Bose A. Photoferrotrophs produce a PioAB electron conduit for extracellular electron uptake. mBio. 2019;10:e02668–19.
Guzman MS, Rengasamy K, Binkley MM, Jones C, Ranaivoarisoa TO, Singh R, et al. Phototrophic extracellular electron uptake is linked to carbon dioxide fixation in the bacterium Rhodopseudomonas palustris. Nat Commun. 2019;10:1355.
Gupta D, Guzman MS, Bose A. Draft genome sequence of a marine photoferrotrophic bacterium, Rhodovulum robiginosum DSM 12329(T). Microbiol Resour Announc. 2019;8:e01684–18.
Straub KL, Rainey FA, Widdel F. Rhodovulum iodosum sp. nov. and Rhodovulum robiginosum sp. nov., two new marine phototrophic ferrous-iron-oxidizing purple bacteria. Int J Sys Evol Microbiol. 1999;49:729–35.
Guzman MS, McGinley B, Santiago-Merced N, Gupta D, Bose A. Draft genome sequences of three closely related isolates of the purple nonsulfur bacterium Rhodovulum sulfidophilum. Genome Announc. 2017;5:e00029–17.
Jiao Y, Newman DK. The pio operon is essential for phototrophic Fe (II) oxidation in Rhodopseudomonas palustris TIE-1. J Bacteriol. 2007;189:1765–73.
Rengasamy K, Ranaivoarisoa T, Singh R, Bose A. An insoluble iron complex coated cathode enhances direct electron uptake by Rhodopseudomonas palustris TIE-1. Bioelectrochemistry. 2018;122:164–73.
Penfold RJ, Pemberton JM. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene. 1992;118:145–6.
Riedel T, Rohlfs M, Buchholz I, Wagner-Dobler I, Reck M. Complete sequence of the suicide vector pJP5603. Plasmid. 2013;69:104–7.
Bose A, Gardel EJ, Vidoudez C, Parra EA, Girguis PR. Electron uptake by iron-oxidizing phototrophic bacteria. Nat Commun. 2014;5:3391.
Poulton RRS. The low-temperature geochemical cycle of iron: From continental fluxes to marine sediment deposition. Am J Sci. 2002;302:774–805.
Kump LR, Seyfried WE. Hydrothermal Fe fluxes during the Precambrian: effect of low oceanic sulfate concentrations and low hydrostatic pressure on the composition of black smokers. Earth Planet Sci Lett. 2005;235:654–62.
Brock TD, Od’ea K. Amorphous ferrous sulfide as a reducing agent for culture of anaerobes. Appl Environ Microbiol. 1977;33:254–6.
Velthuys BR. Electron-dependent competition between plastoquinone and inhibitors for binding to photosystem II. FEBS Lett. 1981;126:277–81.
Wraight CA. Oxidation-reduction physical chemistry of the acceptor quinone complex in bacterial photosynthetic reaction centers: Evidence for a new model of herbicide activity. Isr J Chem. 1981;21:348–54.
Kyle DJ. The 32000 dalton qb protein of photosystem II. Photochem Photobio. 1985;41:107–16.
Michel H, Epp O, Deisenhofer J. Pigment-protein interactions in the photosynthetic reaction centre from Rhodopseudomonas viridis. EMBO J. 1986;5:2445–51.
Oettmeier W. Herbicide resistance and supersensitivity in photosystem II. Cell Mol Life Sci. 1999;55:1255–77.
Trebst A. Inhibitors in the functional dissection of the photosynthetic electron transport system. Photosynth Res. 2007;92:217–24.
Broser M, Glockner C, Gabdulkhakov A, Guskov A, Buchta J, Kern J, et al. Structural basis of cyanobacterial photosystem II Inhibition by the herbicide terbutryn. J Biol Chem. 2011;286:15964–72.
White D. The physiology and biochemistry of prokaryotes. Oxford: Oxford Press; 2007. pp. 112–7.
Heytler PG. Uncoupling of oxidative phosphorylation by carbonyl cyanide phenylhydrazones. I. Some characteristics of m-Cl-CCP action on mitochondria and chloroplasts. Biochemistry. 1963;2:357–61.
Herter SM, Kortluke CM, Drews G. Complex I of Rhodobacter capsulatus and its role in reverted electron transport. Arch Microbiol. 1998;169:98–105.
Spero MA, Brickner JR, Mollet JT, Pisithkul T, Amador-Noguez D, Donohue TJ. Different functions of phylogenetically distinct bacterial complex I isozymes. J Bacteriol. 2016;198:1268–80.
Dupuis A, Darrouzet E, Duborjal H, Pierrard B, Chevallet M, van Belzen R, et al. Distal genes of the nuo operon of Rhodobacter capsulatus equivalent to the mitochondrial ND subunits are all essential for the biogenesis of the respiratory NADH-ubiquinone oxidoreductase. Mol Microbiol. 1998;28:531–41.
Palmer G, Horgan DJ, Tisdale H, Singer TP, Beinert H. Studies on the respiratory chain-linked reduced nicotinamide adenine dinucleotide dehydrogenase. XIV. Location of the sites of inhibition of rotenone, barbiturates, and piericidin by means of electron paramagnetic resonance spectroscopy. J Biol Chem. 1968;243:844–847.
Knaff DB. The cytochrome bc1 complexes of photosynthetic purple bacteria. Photosyn Res. 1993;35:117–33.
Trumpower BL. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–29.
Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiol (Read Enl). 2008;154:3609–23.
Gibson JL, Tabita FR. Nucleotide sequence and functional analysis of cbbR, a positive regulator of the Calvin cycle operons of Rhodobacter sphaeroides. J Bacteriol. 1993;175:5778–84.
van Keulen G, Girbal L, van den Bergh ER, Dijkhuizen L, Meijer WG. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J Bacteriol. 1998;180:1411–1417.
Joshi GS, Zianni M, Bobst CE, Tabita FR. Regulatory twist and synergistic role of metabolic coinducer- and response regulator-mediated CbbR-cbbI interactions in Rhodopseudomonas palustris CGA010. J Bacteriol. 2013;195:1381–1388.
Hartmann T, Leimkuhler S. The oxygen-tolerant and NAD+-dependent formate dehydrogenase from Rhodobacter capsulatus is able to catalyze the reduction of CO2 to formate. FEBS J. 2013;280:6083–96.
Bassegoda A, Madden C, Wakerley DW, Reisner E, Hirst J. Reversible interconversion of CO2 and formate by a molybdenum-containing formate dehydrogenase. J Am Chem Soc. 2014;136:15473–15476.
Lemaire ON, Jespersen M, Wagner T. CO2-Fixation strategies in energy extremophiles: What can we learn from acetogens? Front Microbiol. 2020;11:486.
Deutzmann JS, Sahin M, Spormann AM. Extracellular enzymes facilitate electron uptake in biocorrosion and bioelectrosynthesis. MBio. 2015;6:e00496–00415.
Rotaru AE, Shrestha PM, Liu F, Ueki T, Nevin K, Summers ZM, et al. Interspecies electron transfer via hydrogen and formate rather than direct electrical connections in cocultures of Pelobacter carbinolicus and Geobacter sulfurreducens. Appl Environ Microbiol. 2012;78:7645–51.
Potter M, Steinbuchel A. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromol. 2005;6:552–60.
Ekici S, Pawlik G, Lohmeyer E, Koch HG, Daldal F. Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biochim Biophys Acta. 2012;1817:898–910.
Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–413.
Pettigrew GW, Echalier A, Pauleta SR. Structure and mechanism in the bacterial dihaem cytochrome c peroxidases. J Inorg Biochem. 2006;100:551–67.
Shi L, Dong H, Reguera G, Beyenal H, Lu A, Liu J, et al. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol. 2016;14:651–62.
Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev. 2009;73:155–77.
Singer E, Emerson D, Webb EA, Barco RA, Kuenen JG, Nelson WC, et al. Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS One. 2011;6:e25386.
Wang Z, Leary DH, Malanoski AP, Li RW, Hervey WJ 4th, Eddie BJ, et al. A previously uncharacterized, nonphotosynthetic member of the Chromatiaceae is the primary CO2-fixing constituent in a self-regenerating biocathode. Appl Environ Microbiol. 2015;81:699–712.
Gupta D, Guzman MS, Bose A. Extracellular electron uptake by autotrophic microbes: physiological, ecological, and evolutionary implications. J Ind Microbiol Biotechnol. 2020;47:863–867.
Garber AI, Nealson KH, Okamoto A, McAllister SM, Chan CS, Barco RA, et al. FeGenie: A comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front Microbiol. 2020;11:37.
Thony-Meyer L. Haem-polypeptide interactions during cytochrome c maturation. Biochim Biophys Acta. 2000;1459:316–324.
Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 2010;26:2811–2817.
Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Prokaryotic sulfur oxidation. Curr Opin Microbiol. 2005;8:253–259.
De Smet L, Savvides SN, Van Horen E, Pettigrew G, Van, Beeumen JJ. Structural and mutagenesis studies on the cytochrome c peroxidase from Rhodobacter capsulatus provide new insights into structure-function relationships of bacterial di-heme peroxidases. J Biol Chem. 2006;281:4371–4379.
Alric J, Tsukatani Y, Yoshida M, Matsuura K, Shimada K, Hienerwadel R, et al. Structural and functional characterization of the unusual triheme cytochrome bound to the reaction center of Rhodovulum sulfidophilum. J Biol Chem. 2004;279:26090–26097.
Eichorst SA, Trojan D, Roux S, Herbold C, Rattei T, Woebken D. Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ Microbiol. 2018;20:1041–1063.
Srinivas A, Kumar BV, Sree BD, Tushar L, Sasikala C, Ramana CV. Rhodovulum salis sp. nov. and Rhodovulum viride sp. nov., phototrophic Alphaproteobacteria isolated from marine habitats. Int J Syst Evol Microbiol. 2014;64:957–962.
Kumar PA, Aparna P, Srinivas TN, Sasikala C. Ramana ChV. Rhodovulum kholense sp. nov. Int J Syst Evol Microbiol. 2008;58:1723–1726.
Buchan A, González JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–77.
Prosser JI, Head IM, Stein LY. The family Nitrosomonadaceae, the prokaryotes: alphaproteobacteria and betaproteobacteria. Berlin/Heidelberg: Springer; 2014. pp. 901–18.
Ehrenreich A, Widdel F. Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl Environ Microbiol. 1994;60:4517–4526.
Kappler A, Newman DK. Formation of Fe (III)-minerals by Fe (II)-oxidizing photoautotrophic bacteria. Geochim Cosmochim Acta. 2004;68:1217–26.
Yarzabal A, Brasseur G, Ratouchniak J, Lund K, Lemesle-Meunier D, DeMoss JA, et al. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J Bacteriol. 2002;184:313–17.
Liu J, Wang Z, Belchik SM, Edwards MJ, Liu C, Kennedy DW, et al. Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe (II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front Microbiol. 2012;3:37.
Pantke C, Obst M, Benzerara K, Morin G, Ona-Nguema G, Dippon U, et al. Green rust formation during Fe(II) oxidation by the nitrate-reducing Acidovorax sp. strain BoFeN1. Environ Sci Technol. 2012;46:1439–46.
Etique M, Jorand FPA, Zegeye A, Grégoire B, Despas C, Ruby C. Abiotic process for Fe(II) oxidation and green rust mineralization driven by a heterotrophic nitrate reducing bacteria (Klebsiella mobilis). Environ Sci Technol. 2014;48:3742–51.
Nordhoff M, Tominski C, Halama M, Byrne JM, Obst M, Kleindienst S, et al. Insights into nitrate-reducing Fe(II) oxidation mechanisms through analysis of cell-mineral associations, cell encrustation, and mineralogy in the chemolithoautotrophic enrichment culture KS. Appl Environ Microbiol. 2017;83:e00752–17.
Canfield DE, Jørgensen BB, Fossing H, Glud R, Gundersen J, Ramsing NB, et al. Pathways of organic carbon oxidation in three continental margin sediments. Mar Geol. 1993;113:27–40.
Ouchane S, Agalidis I, Astier C. Natural resistance to inhibitors of the ubiquinol cytochrome c oxidoreductase of Rubrivivax gelatinosus: sequence and functional analysis of the cytochrome bc(1) complex. J Bacteriol. 2002;184:3815–22.
Brasseur G, Saribas AS, Daldal F. A compilation of mutations located in the cytochrome b subunit of the bacterial and mitochondrial bc1 complex. Biochim Biophys Acta. 1996;1275:61–9.
Crofts AR, Berry EA. Structure and function of the cytochrome bc1 complex of mitochondria and photosynthetic bacteria. Curr Opin Struct Biol. 1998;8:501–9.
Karthikeyan R, Singh R, Bose A. Microbial electron uptake in microbial electrosynthesis: a mini-review. J Ind Microbiol Biotechnol. 2019;46:1419–26.
Acknowledgements
The authors acknowledge Prof. Robert Kranz [Washington University in St. Louis (WUSTL)] for valuable assistance with heme protein characterization. We thank the Washington University’s Genome Technology and Access Center (GTAC) for technical guidance and transcriptome sequencing. The authors thank the Institute of Materials Science and Engineering at Washington University in St. Louis, which provided facilities, resources, and assistance in microfabrication. We thank Bradley Evans and Shin-Cheng Tzeng from Proteomics and Mass Spectrometry facility, DDPSC. This work was supported by the following grants to AB: The David and Lucile Packard Foundation Fellowship (201563111), the U.S. Department of Energy (grant number DESC0014613), and the U.S. Department of Defense, Army Research Office (grant number W911NF-18-1-0037). Gordon and Betty Moore Foundation, National Science Foundation (Grant Number 2021822), and the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DEAC5207NA27344 (LLNL-JRNL-812309), and AB was also funded by a Collaboration Initiation Grant, an Office of the Vice-Chancellor of Research Grant, and an International Center for Energy, Environment, and Sustainability Grant from Washington University in St. Louis. EJD is supported by an Institutional Training Grant in Genomic Science from the NIH (T32 HG000045-18). MSG was supported by the Initiative for Maximizing Student Development (IMSD) training grant from the U.S. National Institutes of Health (grant number R25-GM103757). This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344 (LLNL-JRNL-821835).
Author information
Authors and Affiliations
Contributions
DG, MSG, KR, RS, EJD, and AB designed the research. AS, KR, and JMM designed, fabricated, and operated the µ-BEC arrays. DG, MSG, KR, TOR, RS, WB, and BM collected the data. DG, MSG, KR, TOR, RS, EJD, and AB analyzed and interpreted the data. DG, MSG, KR, RS, and AB wrote the manuscript with contributions from other authors. All authors reviewed, revised, and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Gupta, D., Guzman, M.S., Rengasamy, K. et al. Photoferrotrophy and phototrophic extracellular electron uptake is common in the marine anoxygenic phototroph Rhodovulum sulfidophilum. ISME J 15, 3384–3398 (2021). https://doi.org/10.1038/s41396-021-01015-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01015-8