Abstract
Herein, we describe a novel integrated biosensor for performing dielectric spectroscopy to analyze biological samples. We analyzed biomolecule samples with different concentrations and demonstrated that the solution's impedance is highly correlated with the concentration, indicating that it may be possible to use this sensor as a concentration sensor. In contrast with standard spectrophotometers, this sensor offers a low-cost and purely electrical solution for the quantitative analysis of biomolecule solutions. In addition to determining concentrations, we found that the sample solution impedance is highly correlated with the length of the DNA fragments, indicating that the sizes of PCR products could be validated with an integrated chip-based, sample-friendly system within a few minutes. The system could be the basis of a rapid, low-cost platform for DNA characterization with broad applications in cancer and genetic disease research.
Similar content being viewed by others
Introduction
Most conventional DNA detection methods rely on fluorescent labels or dyes1 for data readout or imaging2. Pre-detection sample treatments are required to attach visible markers to analytes to confirm the existence of the target3. Indeed, many of today's mainstream commercial products still use the brightness of a sample's fluorescence emission to quantify DNA fragments4. This relative quantification methodology is commonly limited to indicating whether the value is greater or less than a certain level. In general, fluorescent label-based techniques not only demand highly precise and expensive instrumentation but also introduce unpredictable interference into the detection system, which can lead to inaccurate results5.
Electrochemical sensors, which detect and measure electrical signals instead of fluorescence output as in optical sensors, allow label-free imaging and detection6,7. For example, a highly sensitive semiconducting nanowire sensor that is capable of achieving label-free detection of antibodies at concentrations less than 100 femtomolar has been reported by Stern8. A variety of electrochemical methods based on integrated devices have been employed for label-free DNA detection. These methods are based on technologies such as charge transfer sensors (also known as ion-sensitive field-effect transistor-based sensors)9,10,11, capacitance-based sensors12,13 and impedance-based sensors14,15,16. Impedance measurement, which has been reported as a next-generation imaging technique, is emerging as a powerful tool for biological sensing17,18. However, the development and use of integrated sensors for DNA detection are limited by the need for the immobilization of molecules on the electrodes. The electrode material has to be biologically compatible, which requires additional processes when preparing integrated chips19. Furthermore, because of the nature of the binding and immobilization of targets on the probes, these chips can only be used once and the biological samples cannot be reused.
Here, we measured the bulk electrical properties of DNA solutions. This method overcomes the limitations of affinity-based sensors and enables label-free detection based on an integrated chip. The chip was a 16 × 12 sensor array fabricated using 0.35 μm standard CMOS technology and was designed to perform dielectric spectroscopy (see Supplementary Fig. 1 online). Without any molecular immobilization, the chip was able to detect the impedance changes of suspended DNA samples with different concentrations. This chip was also able to monitor the DNA digestion progress, which is important for some sensitive applications that require eliminating DNA from RNA, such as RT-PCR (reverse transcriptase polymerase chain reaction)20. Furthermore, a series of PCR product measurements demonstrated that the size of the DNA fragments can also be verified using this methodology. Most importantly, the chip is reusable and there is no denaturation of the analyzed biological samples. Thus, the samples can be reused, which is important when analyzing precious or scarce samples. We report a system that utilizes integrated chip-based impedance measurements for the characterization of suspended DNA, including PCR products.
Results
Measurement system
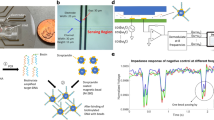
We used a pre-designed 16 × 12 micro-array chip that was fabricated using 0.35-μm CMOS technology (Fig. 1a and Supplementary Fig. 1 online). The pixels are located in the center of the chip and are surrounded by readout and amplifier circuits. During the measurement, the sensing area was immersed in the biological sample solution. The surrounding circuitry was protected from short circuiting by a layer of wax (see Supplementary Fig. 2 online). For the impedance measurements, an AC voltage stimulus was provided to one of the electrodes and the resulting current signal was detected and collected by the other electrode. In this design, all the pixels inside the sensing area share a common electrode to provide the AC signal to the analyte (electrode in Fig. 1b, 1c). Within one pixel cell, the sensing electrode (electrode in Fig. 1b, 1c) was surrounded by the common electrode. Two electrodes were arranged on a flat surface and separated by 1-μm-high and 25-μm-wide insulation barriers (Fig. 1b). In contrast with the parallel electrode architecture described in the literature21,22, which measures the direct (as opposed to fringing) capacitance, the planar electrodes used in this work, take advantage of the fringing field capacitance in the solution (see Supplementary Note online). The sensing electrode was connected to the surrounding circuits. A current-mode transimpedance amplifier, which was located at the end of each column, converted the current signal into a voltage using an off-chip 100 kΩ resistor (Fig. 1c). Finally, the on-chip readout circuit output the converted signals. In this work, we used a function generator (Agilent 33250A) to provide the input signal and an oscilloscope (Agilent Infiniium 54830 DSO) to record both the input (from the function generator) and output (from the sensor chip) signals (see Supplementary Fig. 3 online). Matlab, a technical computing program, was used to analyze the recorded data and calculate the impedance of each sample (see Supplementary Method online).
The micro-array-based biosensor allows the rapid, non-invasive measurement of the impedance of biological solutions.
(a) Photographs of the integrated chip, the sensing area and the excitation circuits. (b) Sensor surface profile and cross section diagram. (c) Schematic diagram of a sensing pixel and the dimensions of each pixel.
This CMOS-based impedance measurement system was fully calibrated using a commercial impedance analyzer (Solartron SI 1260) with NaCl solutions of known concentrations. NaCl solutions are pure resistive solutions and were used to simulate the equivalent circuit for impedance measurements (see Supplementary Note online).
Concentration tests
We used this biochip-based system to measure the impedance of DNA solutions with different concentrations. Concentrated single-stranded DNA fragments from herring sperm with a uniform length of approximately 700 base pairs were purchased from SIGMA-ALDRICH (D7290). This sample comprised DNA molecules without any additional buffer. The molecules were dissolved in deionized (DI) water and diluted to six different concentrations: 12 ng/μl, 30 ng/μl, 60 ng/μl, 120 ng/μl, 240 ng/μl and 600 ng/μl. All the samples were validated using a commercial spectrophotometer (Nanodrop™ 2000). Figure 2a depicts the impedance value of each sample in Cole-Cole plot when the stimulus frequency ranged from 10 kHz to 100 kHz. This frequency range could reflect the electrolyte's characteristics (see Supplementary Note online). The curves represent the six different concentrations and the general impedance of each sample increased gradually as the ssDNA concentration decreased (from left to right). Higher impedance indicates that the solution has lower conductivity. For polyelectrolyte solutions, the conductivity is determined by the properties of the free charge carriers and depend on variables such as the concentration, mobility and number of charges for each carrier23. Biological samples were prepared in DI water and thus, increasing the ssDNA concentration introduced more charge carriers into the electrolyte solution. DNA molecules are negatively charged and the charge is proportional to the number of base pairs24. The concentrations of the different samples were defined in units of mass (ng) over volume (μl). Higher concentrations represent more base pairs per unit volume, resulting in a greater charge density and a lower impedance.
Impedance measurements for DNA and BSA samples with different concentrations.
(a) Cole-Cole plot for the ssDNA samples over the frequency range of 10 kHz to 100 kHz. (b) Cole-Cole plot for the BSA samples over the frequency range of 10 kHz to 100 kHz. (c) Relationship between the biomolecule (ssDNA or BSA) concentration and the solution impedance. The error bars represent the standard deviation of the measurements.
We also analyzed BSA samples (bovine serum albumin, SIGMA-ALDRICH A7906) of different concentrations using the same measurement settings so as to validate the impedance-based method for determining the concentration of biomolecules. Six BSA solutions were prepared: 1 μg/μl, 2 μg/μl, 5 μg/μl, 10 μg/μl, 20 μg/μl and 60 μg/μl. Figure 2b presents the impedance results, which show the same tendency as those of the DNA samples. Solutions with low concentrations had high impedance.
We graphed the impedance data for both the ssDNA and BSA samples for 50 kHz excitation on one plot (Fig. 1c). The impedance values of both samples were continuous and a linear relationship between impedance and concentration was observed in the log-log plot.
Digested DNA tests
After the concentration test, we used the biochip system to perform impedance measurements for a series of digested ssDNA samples. An enzymatic reaction was designed to digest double- and single-stranded DNA into oligo and mononucleotides using DNase I. Under the conditions used, the ssDNA would be completely digested in 15 minutes at room temperature according to the manufacturer's datasheet25,26. The experimental methodology is illustrated in figure 3a. We used ssDNA from herring sperm (240 ng/μl) with a known length range of 587 bp to 831 bp as the analyte. An aliquot of the stock solution (100 μl) was added to each of eight identical tubes containing DNase I and reaction buffer. The reaction time for each tube was precisely controlled by a stopwatch. We stopped the reactions after eight different time intervals - 1 min, 2 min, 4 min, 6 min, 8 min, 14 min, 15 min and 20 min - by adding stop solution and heat inactivating the DNase I. We then performed biochip impedance tests with the samples (Fig. 3b). In general, the impedance of each sample was correlated with the reaction time. Sample number 1, which only underwent a 1-min reaction, had the highest impedance. The impedance gradually decreased as the reaction time increased. For longer reactions, DNase I had more time to cut the DNA molecules, thus resulting in more free charge carriers in the electrolyte. Furthermore, because a smaller molecular size contributes to higher mobility, the solution's conductivity was also boosted by the decrease in the average DNA fragment length with increasing digestion time. The impedances of sample 7 and sample 8 converged, indicating that the reaction had reached completion. The sample impedance was constant after 15 minutes.
PCR product tests
In the DNA digestion test, a longer reaction time resulted in shorter DNA fragments on average and these results therefore suggest that our impedance biochip is able to distinguish DNA fragments of different lengths. A DNA fragment experiment was designed using conventional PCR to produce various DNA fragments of known lengths.
For the PCR, we used the standard plasmid cloning vector pUC19 as the template. We amplified different lengths of double-stranded DNA from the template for use in the biochip measurements. Primers were designed to amplify six different fragment lengths: 148 bp, 309 bp, 964 bp, 1389 bp and 2129 bp (Fig. 4a). Every sample contained the same reaction mixture to help ensure a constant background in the subsequent impedance measurements. The details of the PCR are described in the methods section. The impedance values for samples with different lengths were separated into distinct curves (Figure 4c). Sample 1 was the shortest fragment at 148 bp and had the lowest impedance. In general, the impedance values for the samples increased gradually as the size of the fragment increased. The impedance results for the PCR products followed a tendency similar to that of the DNase I reaction products. These results indicate that the biochip is able to detect differences in the lengths of DNA fragment by measuring the impedance differences. For the PCR products, we also plotted the sample's impedance versus the stimulus frequency (figure 4d) as an alternative way of presenting the impedance data. The impedance for all six samples decreased as the frequency increased. An even more clear relationship was observed when we looked at a specific stimulus frequency. Figure 4e illustrates the linear relationship between the length of the DNA fragments and the measured impedance at 10 kHz.
Impedance measurements for PCR products.
(a) Six different PCR products with different lengths were prepared. (b) The sizes of the PCR products were validated by standard gel electrophoresis. A 2-log DNA ladder from New England BioLabs was used as a reference. The picture was colored using software to make it easier to interpret. (c) Cole-Cole plot showing that the impedance of each sample was dependent on the fragment length. (d) Sample impedance versus the stimulus frequency. (e) The measured impedance versus DNA length in base pairs at 10 kHz.
Discussion
We present an integrated biosensor that is capable of preforming dielectric spectroscopy with biological samples. By detecting the impedance, this novel system is capable of characterizing DNA samples based on their concentration and fragment length. All the label-free experiments were performed using suspended biomolecule solutions without any surface immobilization to probes or bio-functionalization. No further post-fabrication processes are required before taking biological measurements with the integrated CMOS chip. Therefore, the sensor chip system is entirely reusable, which enhances uniformity and reduces the cost of each measurement.
Biological samples can be very precious or scarce and thus, minimizing waste is a key consideration in biosensor design. This micro-array-based design integrates hundreds of micro electrodes and driving circuits into a 5 × 5 mm area. Each measurement requires only 20 μl of sample, as this volume is sufficient to cover the electrodes. Two of the major advantages of this method allow the reuse of the analyte. First, the integrated chip utilizes a non-invasive electrochemical method, dielectric spectroscopy, to analyze the samples. The typical AC stimulus of 500 mVpp is negligible compared with that of gel electrophoresis, which typically requires a supply voltage of more than 100 V DC. Second, the biochip detects the electrical properties of the biological molecules without the need for chemical reactions. The analyzed biological samples are unchanged and can be reused for further analysis.
We investigated the impedances of ssDNA samples with different concentrations using the integrated biosensing system. Current applications of DNA concentration detection systems include the quantification of samples, such as libraries, prior to next-generation sequencing and quantitative PCR. Identical measurements were performed using BSA samples for comparison. Both sets of results show that the solution impedance is related to the biomolecule concentration. Concentrated solutions had lower impedance, indicating higher conductivity. This behaviour can be explained by the role of counter-ions that are attracted by the DNA backbone charges, which are in turn proportional to the concentration. More mobile counter-ions in the vicinity of the DNA results in higher solution conductivity27. On a log-log scale, the solution impedance showed a linear relationship with the sample concentration. This feature makes it possible to use the biochip to determine the concentrations of different samples. In general, the DNA data were clearer and more convincing than the data for the BSA samples. This difference in data quality is most likely due to the molecular structures of the samples. We used short fragments of single-stranded DNA with a length of several hundred base pairs28. Although ssDNA may have some secondary structure due to interactions between its own bases and interactions with other fragments, BSA, as a protein, is expected to have a considerably more complicated structure29,30.
Optical sensors, such as spectrophotometers, which measure the reflection or transmission properties of an analyte as a function of wavelength31, currently dominate the DNA (and other biomolecule) detection sensor market. Most systems employ photometers to convert light signals into electrical signals for further analysis. The complex structure of the optical-electrical system results in reduced accuracy and higher costs. In this work, we report an alternative method for the quantitative analysis of biological samples. Our method significantly reduces the cost by utilizing an integrated sensor. We performed impedance spectroscopy, which measures a sample's ability to conduct electricity as a function of frequency. Furthermore, our system is purely electrical, which reduces the system's complexity and minimizes external interference.
In addition to the DNA concentration, other parameters can be measured using this impedance-based integrated biochip system. We performed a series of impedance measurements for digested ssDNA samples. DNase I was used in this experiment. This enzyme is suitable for eliminating DNA from RNA preparations prior to sensitive applications, such as RT-PCR. The DNase was able to completely digest the ssDNA into oligo- and mononucleotides in a 15-minute reaction at room temperature. We divided the samples into eight groups and the reaction time was precisely controlled. Our results showed that the reaction progress was related to the sample's impedance. The constant solution impedance after 15 minutes indicated that the digestion reaction had reached completion. During the DNA digestion, DNase I randomly cut the DNA strands into smaller pieces. Longer reaction times were expected to result in a shorter average DNA length. The shorter fragments act as charge carriers with higher mobility and thus provide higher solution conductivity. The solution impedance decreased as the digestion reaction progressed until the digestion reached completion at 15 minutes. This experiment demonstrated that the integrated biochip is capable of monitoring the progress of DNA digestion by measuring the impedance.
The DNA digestion experiment also suggested that the length of the DNA fragments is strongly correlated with the solution's conductivity. To validate this relationship, we designed an experiment that used PCR to generate various DNA fragments of known length. We amplified dsDNA, whereas the digestion test used ssDNA. Double-stranded DNA generally has less secondary structure than single-stranded DNA. In addition, the PCR products for each fragment size all had the same DNA sequence, whereas the herring ssDNA would contain a very heterogeneous mixture of DNA sequences. All of these factors minimize the interference in the impedance measurements. As expected, longer fragments were less conductive than shorter fragments. These results indicate that this system is capable of overcoming interference from other molecules in the solution, thus allowing the rapid yet precise quantitative analysis of unpurified PCR products.
The fragment length is one of the most important features of DNA for a wide range of genetic analyses32,33. The correlation between impedance and DNA fragment length observed in this study raises the possibility of using this purely electrical system to validate and characterize PCR products. Gel electrophoresis has dominated this area for more than 40 years. The conventional technique involves imaging the DNA using UV light after the intercalation of ethidium bromide, a highly carcinogenic compound that can also potentially damage the DNA's structure and induce mutations34. Our impedance system is capable of replacing this time-consuming and potentially dangerous technique. The rapid characterization of restriction fragment profiles could potentially be applied to genome assembly, in a manner similar to that of optical mapping35. We present a method for the label-free analysis of suspended samples and we demonstrated that this method can detect differences in the impedance of different PCR products within a few minutes. The samples can even be reused without any further treatment.
The measurement resolution of the DNA parameters (for example, concentration and fragment length) is ultimately limited by system noise. A discussion of noise sources and approaches to enhance measurement resolution is given in Supplementary Note online.
In the work presented herein, a powerful method for DNA characterization was assessed. This method uses an integrated biochip to measure the impedance of different samples. This purely electrical system has demonstrated potential for use in DNA concentration measurements, the analysis of DNA digestion reactions and PCR product validation. All the measurements are performed using suspended samples without any functionalization of the sensor surface. However, to achieve high-accuracy measurements, further systematic calibration is needed. This work opens up the possibility of developing a low-cost, rapid platform for the characterization of DNA and other biological molecules.
Methods
Testing system setup
The micro-array chip was designed using the Cadence Analogue IC design package and the prototype was fabricated using low-cost 0.35 μm CMOS technology by TSMC (Taiwan). The details of the circuit design and functionalities are explained in the Supplementary information. The chip was mounted on PCB and connected to external instruments. A Keithley 4200 system was used to apply biases of 3.3 V DC and 2 V DC and 2 × 50 μA current sources to prevent the circuit from overloading and for signal amplification, respectively. A pre-programmed FPGA (Altera, APEX 20 K) drove the circuit to activate the address of each column of the array. A function generator (Agilent, 33250A 80 MHz) generated 500 mVpp AC signals for the measurements with variable frequency (typically from 10 kHz to 100 kHz). The signals were recorded using an oscilloscope (Agilent, Infiniium 54833A) and impedance data extraction was performed using Matlab (MathWorks). A schematic diagram of the system is shown in Supplementary Figure 3 online.
Preparation of DNA samples with different concentrations
Herring sperm DNA was ordered from SIGMA-ALDRICH (D7290). The single-stranded DNA fragments ranged in size from 587 to 831 base pairs. This DNA was provided as a ready-to-use concentrated solution (9–12 mg/ml DNA). According to the supplier's instructions, we boiled the solution for 10 minutes and then cooled it on ice for another 10 minutes to reduce the likelihood of reannealing of the fragments. We first diluted 10 μl of the concentrated DNA sample with 90 μl of DI water to prepare the stock solution. The concentration of the stock solution was determined to be 1.2 μg/μl (1200 ng/μl) using a Nanodrop™ 2000. We diluted the stock solution with DI water to give different concentrations: 20 μl of stock solution and 20 μl of DI water for a concentration of 600 ng/μl; 10 μl of stock solution and 40 μl of DI water for a concentration of 240 ng/μl; 10 μl of stock solution and 90 μl of DI water for a concentration of 120 ng/μl; 10 μl of stock solution and 190 μl of DI water for a concentration of 60 ng/μl; 5 μl of stock solution and 195 μl of DI water for a concentration of 30 ng/μl; and 2 μl of stock solution and 198 μl of DI water for a concentration of 12 ng/μl. The concentrations of all diluted samples were validated using the Nanodrop and the measured values were the same as the expected values. All of the sample preparation steps were performed at room temperature.
Preparation of BSA samples with different concentrations
BSA was purchased from SIGMA-ALDRICH (MFCD00130384) and was obtained as a white powder. First, 60 grams of BSA powder was dissolved in 1 liter of DI water to prepare a stock solution with a concentration of 60 μg/μl. We diluted the stock solution with DI water to give different concentrations: 20 μl of stock solution and 40 μl of DI water for a concentration of 20 μg/μl; 10 μl of stock solution and 50 μl of DI water for a concentration of 10 μg/μl; 5 μl of stock solution and 55 μl of DI water for a concentration of 5 μg/μl; 2 μl of stock solution and 58 μl of DI water for a concentration of 2 μg/μl; and 2 μl of stock solution and 118 μl of DI water for a concentration of 1 μg/μl.
Preparation DNA digestion time course samples
We prepared eight identical solutions containing 2 μl of DNA (240 ng/μl), 10 μl of reaction buffer, 2 μl of DNase I and 86 μl of DI water. The DNA was digested with DNase I at room temperature and the time was recorded with a timer. After the desired reaction time, we stopped the reaction by adding a stop solution and then heated the sample to 70°C for 10 minutes. We used the same herring sperm ssDNA as that used in the DNA concentration experiment (SIGMA-ALDRICH, D7290). The reaction buffer (Catalog Number: R6273), stop solution (S4809) and DNase I (D5307) were ordered as a reaction package from SIGMA-ALDRICH (AMPD1-1KT).
PCR to produce DNA fragments of different lengths
We used pUC19 as the PCR template and designed primers to amplify double-stranded DNA fragments of different lengths. For the 148 bp fragment, the forward primer was 5′- TATCCGCTCACAATTCCACA-3′ and the reverse primer was 5′- ATTAATGCAGCTGGCACGA-3′. For the 309 bp fragment, the forward primer was 5′-CGCAGGAAAGAACATGTGAG-3′ and the reverse primer was 5′-CGAACGACCTACACCGAACT-3′. For the 501 bp fragment, the forward primer was 5′-GCTTAACTATGCGGCATCAGA-3′ and the reverse primer was 5′-ATTAATGCAGCTGGCACGA-3′. For the 964 bp fragment, the forward primer was 5′-GCTTAACTATGCGGCATCAGA-3′ and the reverse primer was 5′-CGAACGACCTACACCGAACT-3′. For the 1389 bp fragment, the forward primer was 5′-GCTTAACTATGCGGCATCAGA-3′ and the reverse primer was 5′-TGACCAAAATCCCTTAACGTG-3′. For the 2129 bp fragment, the forward primer was 5′-GCTTAACTATGCGGCATCAGA-3′ and the reverse primer was 5′-GCTATGTGGCGCGGTATTAT-3′. All the primers were ordered from Eurofins UK.
For each 50 μl reaction, we added 34.5 μl of DI water, 5 μl of buffer, 2 μl of dNTP mix (5 mM), 2 μl of MgCl2 solution (2 mM), 0.5 μl of DNA polymerase, 2.5 μl of forward primer, 2.5 μl of reverse primer and 1 μl of plasmid. The pUC19 plasmid, BIOTAQ™ polymerase, reaction buffer, MgCl2 solution and dNTPs were all from Bioline. The PCRs were performed using a GS1 G-Storm thermocycler.
References
Froehling, P. Dendrimers and dyes—a review. Dyes Pigm. 48, 187–195 (2001).
Chaji, G., Nathan, A. & Pankhurst, Q. Merged phototransistor pixel with enhanced near infrared response and flicker noise reduction for biomolecular imaging. Appl. Phys. Lett. 3–5 (2008).
Ramsay, G. DNA chips: state-of-the art. Nat. Biotechnol. (1998).
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–45 (2008).
Evanko, D. Label-free microscopy. Nat. Methods 7, 36–36 (2010).
Sadik, O. A., Aluoch, A. O. & Zhou, A. Status of biomolecular recognition using electrochemical techniques. Biosens. Bioelectron. 24, 2749–65 (2009).
Drummond, T. G., Hill, M. G. & Barton, J. K. Electrochemical DNA sensors. Nat. Biotechnol. 21, 1192–9 (2003).
Stern, E. et al. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature 445, 519–22 (2007).
Maruyama, Y., Terao, S. & Sawada, K. Label free CMOS DNA image sensor based on the charge transfer technique. Biosens. Bioelectron. 24, 3108–12 (2009).
Rothberg, J. M. et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–52 (2011).
Gonçalves, D., Prazeres, D. M. F., Chu, V. & Conde, J. P. Label-free electronic detection of biomolecules using a-Si:H field-effect devices. J. Non-Cryst. Solids 352, 2007–2010 (2006).
Stagni, C. et al. A Fully Electronic Label-Free DNA Sensor Chip. Sensors (Peterborough, NH) 7, 577–585 (2007).
Barbaro, M. et al. A CMOS, fully integrated sensor for electronic detection of DNA hybridization. IEEE Electron Device Lett. 27, 595–597 (2006).
Manickam, A., Chevalier, A., Mcdermott, M., Ellington, A. D. & Hassibi, A. A CMOS Electrochemical Impedance Spectroscopy (EIS) Biosensor Array. IEEE Trans. Biomed. Circuits Syst. 4, 379–390 (2010).
Yufera, A. & Rueda, A. A CMOS bio-impedance measurement system. IEEE DDECS (2009)
Sun, T. et al. On-chip epithelial barrier function assays using electrical impedance spectroscopy. Lab Chip 10, 1611–7 (2010).
Wang, W. et al. Single cells and intracellular processes studied by a plasmonic-based electrochemical impedance microscopy. Nat. Chem. 3, 249–255 (2011).
Doerr, A. Imaging impedance. Nat. Methods 8, 202–202 (2011).
Daniels, J. S. & Pourmand, N. Label-Free Impedance Biosensors: Opportunities and Challenges. Electroanalysis 19, 1239–1257 (2007).
Sonza, S., Mutimer, H. & Deacon, N. Pitfalls of processed pseudogenes in RT-PCR. Bio Techniques 24, 585–588 (1998).
Yi, M., Jeong, K.-H. & Lee, L. P. Theoretical and experimental study towards a nanogap dielectric biosensor. Biosens. Bioelectron. 20, 1320–6 (2005).
Mannoor, M. S., James, T., Ivanov, D. V., Beadling, L. & Braunlin, W. Nanogap dielectric spectroscopy for aptamer-based protein detection. Biophys J. 98, 724–32 (2010).
Bockris, J. O. M. & Reddy, A. K. N. Mod. Electrochem. (1998).
Brody, J. R. & Kern, S. E. History and principles of conductive media for standard DNA electrophoresis. Anal.Biochem. (2004).
Huang, Z., Fasco, M. & Kaminsky, L. Optimization of DNase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. Bio Techniques 20, 1012–1020 (1996).
Sanyal, A., O'Druscikk, S. W., Bolander, M. E. & Sarkar, G. An effective method of completely removing contaminating genomic DNA from an RNA sample to be used for PCR. Mol. Biotechnol. 8, 135–137 (1997).
Chang, H. et al. DNA counterion current and saturation examined by a MEMS-based solid state nanopore sensor. Biomed. Microdevices 8, 263–9 (2006).
Trinh, T. & Subdeb, R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E.coli. Nature 352, 544–547 (1991).
Anfinsen, C. Principles that Govern the Folding of Protein Chains. Science 181, 223–230 (1973).
Li, H., Helling, R., Tang, C. & Wingreen, N. Emergence of preferred structures in a simple model of protein folding. Science (New York, N.Y.) 666–9 (1996).
Kamentsky, L., Melamed, M. & Derman, H. Spectrophotometer: New Instrument for Ultrarapid Cell Analysis. Science 150, 630–631 (1965).
Volkmuth, W. & Austin, R. DNA electrophoresis in microlithographic arrays. Nature (1992).
McHardy, A., Martín, H. & Tsirigos, A. Accurate phylogenetic classification of variable-length DNA fragments. Nature 4, 63–72 (2006).
Hirschman, S. Z. Inhibitors of DNA Polymerases of Murine Leukemia Viruses: Activity of Ethidium Bromide. Science 173, 441–443 (1971).
Kidd, J. M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008).
Acknowledgements
We thank Dr. Ashwin Seshia (University of Cambridge, UK) for the use of laboratory facilities and Richard Bayford and Martina Callaghan (Middlesex University, UK) for helpful discussions about the impedance measurements.
Author information
Authors and Affiliations
Contributions
R.C. and A.N. designed the CMOS chip; H.M., Y.S. and A.N. designed the concentration and digestion experiments; H.M. and Y.S. prepared the samples for the concentration and digestion experiments; H.M., R.W., C.J. and A.N. designed the PCR product experiments; H.M. and R.W. designed and performed the PCR and gel electrophoresis steps; H.M. performed the biochip-based measurements and data extraction; and H.M. and J.L. performed the system calibration and simulation.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Ma, H., Wallbank, R., Chaji, R. et al. An impedance-based integrated biosensor for suspended DNA characterization. Sci Rep 3, 2730 (2013). https://doi.org/10.1038/srep02730
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02730
This article is cited by
-
Longer amplicons provide better sensitivity for electrochemical sensing of viral nucleic acid in water samples using PCB electrodes
Scientific Reports (2022)
-
Multi-frequency impedance sensing for detection and sizing of DNA fragments
Scientific Reports (2021)
-
Ultrasensitive and label-free biosensor for the detection of Plasmodium falciparum histidine-rich protein II in saliva
Scientific Reports (2019)
-
Label-free electrochemical impedance spectroscopy using a micro interdigitated electrode inside a PCR chip for real-time monitoring
Microsystem Technologies (2019)
-
Frequency-dependent conductance change of dielectrophoretic-trapped DNA-labeled microbeads and its application in DNA size determinations
Microfluidics and Nanofluidics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.