Abstract
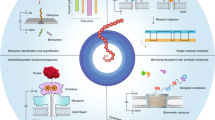
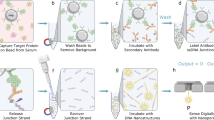
Decoding the identity of biomolecules from trace samples is a longstanding goal in the field of biotechnology. Advances in DNA analysis have substantially affected clinical practice and basic research, but corresponding developments for proteins face challenges due to their relative complexity and our inability to amplify them. Despite progress in methods such as mass spectrometry and mass cytometry, single-molecule protein identification remains a highly challenging objective. Towards this end, we combine DNA nanotechnology with single-molecule force spectroscopy to create a mechanically reconfigurable DNA nanoswitch caliper capable of measuring multiple coordinates on single biomolecules with atomic resolution. Using optical tweezers, we demonstrate absolute distance measurements with ångström-level precision for both DNA and peptides, and using multiplexed magnetic tweezers, we demonstrate quantification of relative abundance in mixed samples. Measuring distances between DNA-labelled residues, we perform single-molecule fingerprinting of synthetic and natural peptides, and show discrimination, within a heterogeneous population, between different posttranslational modifications. DNA nanoswitch calipers are a powerful and accessible tool for characterizing distances within nanoscale complexes that will enable new applications in fields such as single-molecule proteomics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data supporting the findings of this paper are available from the supplementary files and the corresponding authors upon reasonable request. Source data are provided with this paper.
Code availability
Code used to analyse data in this paper is available from the corresponding authors upon reasonable request.
References
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Restrepo-Pérez, L., Joo, C. & Dekker, C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 13, 786–796 (2018).
Callahan, N., Tullman, J., Kelman, Z. & Marino, J. Strategies for development of a next-generation protein sequencing platform. Trends Biochem. Sci. 45, 76–89 (2020).
Timp, W. & Timp, G. Beyond mass spectrometry, the next step in proteomics. Sci. Adv. 6, eaax8978 (2020).
Alfaro, J. A. et al. The emerging landscape of single-molecule protein sequencing technologies. Nat. Methods 18, 604–617 (2021).
Swaminathan, J., Boulgakov, A. A. & Marcotte, E. M. A theoretical justification for single molecule peptide sequencing. PLoS Comput. Biol. 11, e1004080 (2015).
Yao, Y., Docter, M., van Ginkel, J., de Ridder, D. & Joo, C. Single-molecule protein sequencing through fingerprinting: computational assessment. Phys. Biol. 12, 055003 (2015).
Ohayon, S., Girsault, A., Nasser, M., Shen-Orr, S. & Meller, A. Simulation of single-protein nanopore sensing shows feasibility for whole-proteome identification. PLoS Comput. Biol. 15, e1007067 (2019).
van Ginkel, J. et al. Single-molecule peptide fingerprinting. Proc. Natl Acad. Sci. USA 115, 3338 (2018).
Swaminathan, J. et al. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat. Biotechnol. 36, 1076–1082 (2018).
Rosen, C. B., Rodriguez-Larrea, D. & Bayley, H. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat. Biotechnol. 32, 179–181 (2014).
Kennedy, E., Dong, Z., Tennant, C. & Timp, G. Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol. 11, 968–976 (2016).
Restrepo-Pérez, L., Wong, C. H., Maglia, G., Dekker, C. & Joo, C. Label-free detection of post-translational modifications with a nanopore. Nano Lett. 19, 7957–7964 (2019).
Ouldali, H. et al. Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol. 38, 176–181 (2020).
Zhao, Y. et al. Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat. Nanotechnol. 9, 466–473 (2014).
Ohshiro, T. et al. Detection of post-translational modifications in single peptides using electron tunnelling currents. Nat. Nanotechnol. 9, 835–840 (2014).
Koussa, M. A., Halvorsen, K., Ward, A. & Wong, W. P. DNA nanoswitches: a quantitative platform for gel-based biomolecular interaction analysis. Nat. Methods 12, 123–126 (2014).
Halvorsen, K., Schaak, D. & Wong, W. P. Nanoengineering a single-molecule mechanical switch using DNA self-assembly. Nanotechnology 22, 494005 (2011).
Yang, D., Ward, A., Halvorsen, K. & Wong, W. P. Multiplexed single-molecule force spectroscopy using a centrifuge. Nat. Commun. 7, 11026 (2016).
Kim, J., Zhang, C.-Z., Zhang, X. & Springer, T. A. A mechanically stabilized receptor–ligand flex-bond important in the vasculature. Nature 466, 992–995 (2010).
Pfitzner, E. et al. Rigid DNA beams for high-resolution single-molecule mechanics. Angew. Chem. Int. Ed. 52, 7766–7771 (2013).
Kilchherr, F. et al. Single-molecule dissection of stacking forces in DNA. Science 353, aaf5508 (2016).
Kostrz, D. et al. A modular DNA scaffold to study protein–protein interactions at single-molecule resolution. Nat. Nanotechnol. 14, 988–993 (2019).
Gosse, C., Strick, T. R. & Kostrz, D. Molecular scaffolds: when DNA becomes the hardware for single-molecule investigations. Curr. Opin. Chem. Biol. 53, 192–203 (2019).
Ma, X. et al. Interactions between PHD3-bromo of MLL1 and H3K4me3 revealed by single-molecule magnetic tweezers in a parallel DNA circuit. Bioconjug Chem. 30, 2998–3006 (2019).
Hatch, K., Danilowicz, C., Coljee, V. & Prentiss, M. Demonstration that the shear force required to separate short double-stranded DNA does not increase significantly with sequence length for sequences longer than 25 base pairs. Phys. Rev. E. 78, 011920 (2008).
Watson, J. D. & Crick, F. H. C. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
Woodside, M. T. et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl Acad. Sci. USA 103, 6190–6195 (2006).
Bustamante, C., Marko, J. F., Siggia, E. D. & Smith, S. Entropic elasticity of λ-phage DNA. Science 265, 1599–1600 (1994).
Abello, N., Kerstjens, H. A. M., Postma, D. S. & Bischoff, R. Selective acylation of primary amines in peptides and proteins. J. Proteome Res. 6, 4770–4776 (2007).
Zhang, X., Halvorsen, K., Zhang, C.-Z., Wong, W. P. & Springer, T. A. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science 324, 1330–1334 (2009).
Oesterhelt, F. et al. Unfolding pathways of individual bacteriorhodopsins. Science 288, 143–146 (2000).
Carrion-Vazquez, M. et al. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Mol. Biol. 10, 738–743 (2003).
Oda, E. et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288, 1053 (2000).
Czabotar, P. E. et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl Acad. Sci. USA 104, 6217 (2007).
Sattler, M. et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986 (1997).
Apweiler, R. et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 (2004).
Adamczyk, M., Gebler, J. C. & Wu, J. Selective analysis of phosphopeptides within a protein mixture by chemical modification, reversible biotinylation and mass spectrometry. Rapid Commun. Mass Spectrom. 15, 1481–1488 (2001).
Meinhart, A. & Cramer, P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430, 223–226 (2004).
Phatnani, H. P. & Greenleaf, A. L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 (2006).
Kim, M., Suh, H., Cho, E. J. & Buratowski, S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284, 26421–26426 (2009).
Eick, D. & Geyer, M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113, 8456–8490 (2013).
Knight, Z. A. et al. Phosphospecific proteolysis for mapping sites of protein phosphorylation. Nat. Biotechnol. 21, 1047–1054 (2003).
Ribeck, N. & Saleh, O. A. Multiplexed single-molecule measurements with magnetic tweezers. Rev. Sci. Instrum. 79, 094301 (2008).
De Vlaminck, I. et al. Highly parallel magnetic tweezers by targeted DNA tethering. Nano Lett. 11, 5489–5493 (2011).
Cnossen, J. P., Dulin, D. & Dekker, N. H. An optimized software framework for real-time, high-throughput tracking of spherical beads. Rev. Sci. Instrum. 85, 103712 (2014).
Smith, S. B., Finzi, L. & Bustamante, C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science 258, 1122 (1992).
Danilowicz, C., Greenfield, D. & Prentiss, M. Dissociation of ligand–receptor complexes using magnetic tweezers. Anal. Chem. 77, 3023–3028 (2005).
Shon, M. J., Rah, S.-H. & Yoon, T.-Y. Submicrometer elasticity of double-stranded DNA revealed by precision force-extension measurements with magnetic tweezers. Sci. Adv. 5, eaav1697 (2019).
Sen, Y.-H., Jain, T., Aguilar, C. A. & Karnik, R. Enhanced discrimination of DNA molecules in nanofluidic channels through multiple measurements. Lab. Chip 12, 1094–1101 (2012).
Keyser, U. F. et al. Direct force measurements on DNA in a solid-state nanopore. Nat. Phys. 2, 473–477 (2006).
Mulhall, E. M. et al. Single-molecule force spectroscopy reveals the dynamic strength of the hair-cell tip-link connection. Nat. Commun. 12, 849 (2021).
Bustamante, C., Chemla, Y. R. & Moffitt, J. R. High-resolution dual-trap optical tweezers with differential detection: instrument design. Cold Spring Harb. 2009, pdb.ip73 (2009).
Lipfert, J., Hao, X. & Dekker, N. H. Quantitative modeling and optimization of magnetic tweezers. Biophys. J. 96, 5040–5049 (2009).
Dulin, D. et al. High spatiotemporal-resolution magnetic tweezers: calibration and applications for DNA dynamics. Biophys. J. 109, 2113–2125 (2015).
De Vlaminck, I., Henighan, T., van Loenhout, M. T. J., Burnham, D. R. & Dekker, C. Magnetic forces and DNA mechanics in multiplexed magnetic tweezers. PLoS ONE 7, e41432 (2012).
Yu, Z. et al. A force calibration standard for magnetic tweezers. Rev. Sci. Instrum. 85, 123114 (2014).
Acknowledgements
We thank S. Buratowski, M. Bao and all members of the Wong and Shih Laboratories for helpful discussions and comments on the paper. This work was funded by support from ONR award no. N000141510073, Smith Family Foundation Odyssey Award, grant no. NIH NIGMS R35 GM119537 (W.P.W.) and the Wyss Institute at Harvard. E.K. acknowledges support from the Human Frontier Science Program (grant no. LT001077/2015-C).
Author information
Authors and Affiliations
Contributions
W.P.W. and W.M.S. conceived the project. P.S., T.E.T., D.Y., W.P.W. and W.M.S. designed the experiments. J.I.M. conducted experiments to label peptides with the DNA handles. P.S. conducted experiments with dual-trap optical tweezers. D.Y., P.S. and T.E.T. conducted experiments with magnetic tweezers. D.Y., P.S., H.T.B. and W.P.W. performed data analysis. A.W., E.K., S.C., Y.L., B.N. and A.J.-B. contributed to early experiments. All authors discussed the results and analysis and contributed to the paper, with the initial draft written by P.S. and W.P.W.
Corresponding authors
Ethics declarations
Competing interests
W.M.S. and W.P.W. have filed patent applications for various aspects of this work.
Additional information
Peer review information Nature Nanotechnology thanks Chirlmin Joo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10, Note 1 and Tables 1–4.
Rights and permissions
About this article
Cite this article
Shrestha, P., Yang, D., Tomov, T.E. et al. Single-molecule mechanical fingerprinting with DNA nanoswitch calipers. Nat. Nanotechnol. 16, 1362–1370 (2021). https://doi.org/10.1038/s41565-021-00979-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-021-00979-0
This article is cited by
-
Single-molecule force stability of the SARS-CoV-2–ACE2 interface in variants-of-concern
Nature Nanotechnology (2024)
-
Full-length single-molecule protein fingerprinting
Nature Nanotechnology (2024)
-
DNA T-shaped crossover tiles for 2D tessellation and nanoring reconfiguration
Nature Communications (2023)
-
Enzyme-less nanopore detection of post-translational modifications within long polypeptides
Nature Nanotechnology (2023)