Abstract
Our recent research revealed that pinewood nematode (PWN) possesses few genes encoding enzymes for degrading α-pinene, which is the main compound in pine resin. In this study, we examined the role of PWN microbiome in xenobiotics detoxification by metagenomic and bacteria culture analyses. Functional annotation of metagenomes illustrated that benzoate degradation and its related metabolisms may provide the main metabolic pathways for xenobiotics detoxification in the microbiome, which is obviously different from that in PWN that uses cytochrome P450 metabolism as the main pathway for detoxification. The metabolic pathway of degrading α-pinene is complete in microbiome, but incomplete in PWN genome. Experimental analysis demonstrated that most of tested cultivable bacteria can not only survive the stress of 0.4% α-pinene, but also utilize α-pinene as carbon source for their growth. Our results indicate that PWN and its microbiome have established a potentially mutualistic symbiotic relationship with complementary pathways in detoxification metabolism.
Similar content being viewed by others
Introduction
The pinewood nematode (PWN) Bursaphlenchus xylophilus is an important invasive plant parasitic nematode that has caused heavy mortality of pine trees, which is called pine wilt disease (PWD), in introduced regions including Japan, Korea, China and Portugal1,2. Pines are extremely rich in resins containing resin acid, terpenoids (especially α-pinenes), benzoate, phenolic compounds, stilbenoids, etc. and those aromatic compounds play important roles in mediating various plant–herbivore interactions. Living in pine trees, PWNs have to survive the stresses of those compounds. Some of them such as terpenoids are membrane-destructive and could inhibit reproduction and development of the nematodes3,4,5. Nematode activities including feeding cause injuries to pine trees, which cause their parenchyma cells to synthesize more terpenoids, benzoic acid and ethylene thus further deteriorate the living condition of the PWNs6,7,8,9. Consequently, PWNs have to cope with a rather toxic condition within the pine trees. Our recent study of the PWN transcriptomes showed that the main metabolic pathway for xenobiotics detoxification in PWN is cytochrome P450 metabolism and PWN does not have a functional pathway that contains all enzymes needed for terpenoids degradation10, suggesting that PWNs may utilize alternative mechanisms for terpenoids degradation.
In this study, we explored the role of microbiome of PWN in terpenoids degradation by metagenomics methods, trying to explore the mechanism of PWN detoxification from the relationship and interaction of microbe-eukaryotic organisms. Recently, by applying new technologies including metagenomics, most eukaryotic organisms have been found to be intimately associated with a complex community of beneficial microbes, called microbiomes, which can be critical for their development and survival11,12,13,14. Nematodes have developed a variety of association types with their microbiomes and the association between nematodes and their corresponding microbiomes have been extensively investigated15. In particular, entomopathogenic nematodes (Steinernema and Heterorhabditis) and their endosymbiotic bacteria (Xenorhabdus and Photorhabdus) has been studied as a model system for nematode-bacterium symbiosis16,17. Additionally, microbial-eukaryotic lateral gene transfer (LGT) has been documented through transcriptome and genome analysis of plant-parasitic nematodes and some transferring genes are ancient acquisition and prevalent among plant-parasitic nematodes, which play a role in evolution of parasitism15,18,19. A variety of bacteria associated with plant-parasitic nematodes, such as cyst nematodes Heterodera20,21, burrowing nematodes Radophoblus22, Dagger nematodes Xiphinema23, have been reported. Also, previous studies have shown that a number of bacteria were associated with pinewood nematodes, Bursaphelenchus xylophilus24,25,26,27,28,29,30 and B. mucronatus31. Bacteria were obviously observed on the surface of B. xylophilus using a light microscopy, transmission electron microscopy (TEM) and scanning electron microscopy (SEM)32. Some bacterial strains have been isolated and identified by culture-dependent method24,25,26. Recently, more bacteria were identified through sequencing the 16S rRNA gene sequences27,28. Bacterial diversity associated with the Chinese PWN was measured by construction of a fosmid library29 and a 16S rRNA gene library30. Moreover, some bacterial species association with PWN were found even after nematodes were continuously reared in laboratory, indicating a strong specific bacteria–nematode relationship between B. xylophilus and its associated bacteria28. We also found that some bacteria were difficult to be removed from the nematode by surface sterilization (unpublished observation). A mutualistic symbiosis between B. xylophilus and bacteria strains are a reciprocal exchange of nutrients, in which bacteria could help the growth and reproduction of the nematode33,34. Furthermore, it has been hypothesized that the symbiotic bacteria associated with PWN could play a crucial role on pine wilt by producing a phytotoxin called phenylacetic acid (PA)24,25,26,35,36,37,38. However, negative experimental evidence has been presented39. Similar as above, some terpenoids of pine host are also harmful to bacteria associated with the nematode. It was reported that the Douglas fir α-pinene could inhibit the growth of a variety of bacteria and all terpenoids could inhibit bacteria in guts of Douglas fir tussock moth larvae at concentrations normally present in the fir needle diet (0.27 mg/g)40. Therefore, metabolism of xenobiotics detoxification is important to both the phytophagous animals and their symbiotic bacteria.
To examine the role of microbiome in xenobiotics detoxification, which is essential for both bacteria and nematodes survival in their pine host, we have carried out metagenomic analysis of the microbiomes of PWNs. Metagenomics has been a major approach to study the composition, function, evolution of various microbiota and interactions between microbiomes and their hosts11,41,42. We aim to examine the metabolic pathways in microbiome metagenomes and compare to that in the PWN genome for understanding the mechanisms underlying detoxification metabolism. More importantly, we have examined the potential capacity for utilization of α-pinene and benzoate as carbon sources in the cultivable symbionts.
Results
Metagenomic analysis and functional annotation
We obtained 38,750,000 reads (2.91 Gb in total) from genomic DNA harvested from enriched bacteria. After filtering out adapters, low quality reads and PCR duplication, 77.4% reads were available for analysis. These high quality reads were assembled into 99,049 contigs (> 200 bp), totalling 78 Mb. The mean coverage of bases in all contigs is 29 bp (range from 1 – 474 bp). The largest contig is 62,542 bp and the average length is 786 bp (Table 1). A total of 155,722 open reading frames (ORFs) were predicted from assembled contigs, including 24% complete and 76% partial ORFs. The coding percentage is 87.8% of the total contig lengths (Table 1). After group analysis by CD-HIT-EST, we obtained 131,100 non-redundant metagenes. We annotated protein function encoded by these metagenes through BLAST searches. In total, 76,467 metagenes (58.3%) were assigned to 4104 KEGG orthologs (KOs). Among them, metabolisms of amino acid, carbohydrate, xenobiotics biodegradation and lipid were the four top abundant categories (supplemental Table S1). Glycine, serine and threonine metabolism (map00260), butanoate metabolism (map00650), benzoate degradation (map00632) and fatty acid metabolism (map00071) were the top pathway in each category, respectively. The top 24 pathways are shown in Figure 1.
To examine whether the metagenome dataset obtained from the enriched bacteria is biased, we obtained another metagenome dataset by sequencing total genomic DNA of PWN, which was newly collected from a pine host in the same region (Zhejiang Province). Bacterial contigs were then isolated from the genome sequences of PWN. A total of 53,482 bacterial contigs (> 200 bp) with 23 Mb in length were obtained and 22,100 ORFs were predicted from the dataset of directly sequencing metagenome (Table 1). As we expected, the result of KEGG functional annotation is similar to the result obtained from the bacteria enriched metagenome (Figure 1, supplemental table S1). We used the pared data, i.e. the numbers of metagenes involved in each KEGG pathways in the two metagomes (Xi, Yi), to calculate the sample Pearson correlation coefficient and the result showed highly correlation between them (r = 0.943, n = 130). Therefore, the metagenome obtained from enriched bacterial is unbiased.
Taxonomic assignment of bacteria symbiosis with PWN
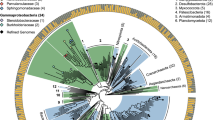
Taxonomic assignment was performed to determine composition of the bacterial consortia in the metagenome. We first matched the reads of the bacterial enriched metagenome to the bacterial genomes deposited in NCBI. To our surprise, only 9.4% reads could directly be matched (mismatches < or = 2 bp in the first 35-bp region, identity > 90%) and almost all of those matched reads were assigned to phylum Proteobacteria (99.8%). This low rate of matched reads is due largely to the stringent requirement by short reads alignment programs (in this case, SOAPaligner), which allow very low number of misalignment. Then, we used the assembled contigs to search against the NCBI bacterial database using BLAST. Altogether 44.9 Mb (57.7%) of assembled contigs with 21,891,219 reads (79.9% of the total) could be matched to the bacterial genomes and had a taxonomic assignment (cut-off 1 × 10−8, identity > 90%). About 95% of mapped reads were assigned to the phylum Proteobacteria, including 46.6% to the Alphaproteobacteria, 38.3% to the Gammaproteobacteria and 10.1% to Betaproteobacteria. The remaining was assigned to the phylum Bacteroidetes (4.4%) and other groups (0.6%). Among them, more than 93% of mapped reads were assigned to nine bacterial families (with a coverage > 1%), in which two families Rhizobiaceae and Xanthomonadaceae occupied 60% of mapped reads (Figure 2). At the genus level, the most common genera of bacteria symbiosis with PWN were Stenotrophomonas, Agrobacterium, Rhizobium, Pseudomonas, Herbaspirillum, Caulobacter, Pedobacter, Novospingobium and Sphingomonas, for each with coverage larger than 1% (supplemental Figure S1A).
The bacterial diversity associated with the PWN strain was also measured by construction of a 16S rRNA gene library30. 26 operational taxonomic units (OTUs) were obtained from 73 clones (with 97% identity cutoff). Of them, four OTUs belonged to Alphaproteobacteria, eight to Betaproteobacteria, ten to Gammaproteobacteria and four to Bacteoidetes. Among them, Stenotrophomonas had the most abundant clones and then followed by Achromobacter, Agrobacterium, Herbaspirillum, Variovorax, Dyella, Pedobacter, Sphingobacterium, Pseudomonas, Novosphingobium, Sphingomonas, etc. (supplemental Figure S1C). Comparison of bacterial compositions obtained through 16S rRNA sequencing from metagenome sequencing showed these two methods revealed similar composition, although richness shows some difference (Figure 2).
Taxonomic assignment of the directly sequencing metagenome showed that less than 20% contigs have a definite taxonomic affiliation. Of them, with more than 70% mapped reads were assigned to Stenotrophomonas genus. So Stenotrophomonas is the most prevalent bacterial strain. Agrobacterium was next and then followed by Achromobacter, Variovorax, Rhizobium and Bordetella. Genus Pseudomonas, Sphingomonas and Pedobacter were also richer than others (supplemental Figure S1C).
Detoxification metabolism of bacteria symbiosis with PWN by metagenome analysis
We are particularly interested in metabolism pathways of xenobiotics biodegradation, because xenobiotics detoxification systems are able to remove compounds from the complex mixture produced by metabolic process or from environment. We found that enzymes involved in metabolism of xenobiotic biodegradation were very abundant in the bacterial metagenome. A total of 12,601 metagenes (16.5%) were involved in this category in the bacteria enriched metagenome. Among them, metagenes encoding enzymes involved in the pathway of benzoate degradation were more abundant than in other pathways (Figure 3), with a complete 3-oxoadipate pathway for benzoate degradation through catechol- the hydroxylation pathway (supplemental Figure S2). Benzoate is a common intermediate in the anaerobic metabolism of aromatic compounds. Toxic compounds, such as phenolic compounds, polycyclic aromatic hydrocarbons, could firstly be degraded to benzoate through some other detoxification pathways and then enter into the pathway of benzoate degradation to be further degraded. The terminal product acetyl-CoA could be utilized in the pathway of citrate cycle. So that benzoate degradation and its related pathways are very important for xenobiotics detoxification in the bacterial consortia symbiosis with PWNs.
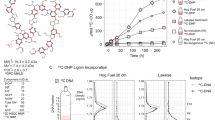
Pinenes are the major components of turpentine which are produced in significant quantities by pine trees. KEGG function annotation of the bacterial metagenome showed that enzymes encoded by 1212 metagenes have activities in metabolism of limonene and pinene degradation (Figure 3). Except for the alpha-pinene oxide lyase (EC 5.5.1.10), which was not found because of no reference (i.e., nucleotide) sequence kept in GenBank, all enzymes involved in the pathway of α-pinene degradation are found in the dataset of bacteria enriched metagenome (Figure 4). The same results were also acquired from the directly sequencing metagenome (Figure 3). Pinenes are monoterpenoid compounds and no pathway of monoterpenoid biosynthesis (map00902) was found in both the bacterial metagenome and the PWN genome, indicating that both the nematode and its symbiotic bacteria could not synthesize monoterpenoid compounds. However, host pine tree can synthesize a large amount of monterpenoids (especially α-pinene) as defensive compounds, to PWN and its symbiotic bacteria, these compounds are toxic and must be degraded by detoxification metabolism. Thus, enzymes involved in monoterpenoid degradation in bacteria metagenome are likely directed at compounds synthesized by the host tree.
Enzymes involved in metabolic pathway of α-pinene degradation in the bacterial metagenomes based on KEGG functional annotation.
Map was downloaded from the KEGG server with our data mapping to the pathway (http://www.kegg.jp/kegg/tool/color_a_pathway.html).
Experimental assessment on the capacity of symbiotic bacteria surviving and utilizing α-pinene and benzoate
We then attempted to identify which bacterium or bacteria could produce enzymes involved in terpenoids degradation. First, we isolated bacterial strains from PWN by culture-dependent method and identified 12 bacterial strains from 188 clones based on 16S rRNA gene sequences. Among them, Stenotrophomonas was the most abundant strain, followed by Cytophaga and Sphingobacterium. Herbaspinillum, Agrobacterium and Pseudomonas were richer than Pedobacter, Achromobacter, Sphingomonas, Variovorax, Ralstonia and Dyella (supplemental Figure S1D).
To detect the capacities of bacteria surviving the stress of α-pinene, we used 10 cultivable bacterial strains to test their potential abilities by culturing them on LB medium containing 0.4% α-pinene, contrasted to those grown on LB medium without α-pinene. The result showed that seven out of ten bacterial strains could grow on LB medium containing α-pinene (Figure 5). Pseudomonas and Cytophaga could grow very well under the stress. Agrobacterium, Stenotrophomonas, Achromobacter, Herbaspirillum and Sphingobacterium showed slow growth at the early stage. The other three, Sphingomonas,Variovorax and Pedobacter, however, could not grow under the stress of 0.4% α-pinene.
Then, we tested the capacity of those strains utilizing α-pinene as carbon source for their growth by culturing them in M9 buffer containing 0.4% α-pinene. Six tolerant strains could also take α-pinene as carbon source for their growth (with OD600 of > 0.01 in 24 h), except Sphingobacterium. Among them, Pseudomonas could grow much better than others in the M9 buffer containing 0.4% α-pinene (Figure 6A). It means that Pseudomonas is more efficiently utilizing α-pinene as the carbon source for its growth.
Moreover, we tested the capacity of those strains utilizing benzoate as the carbon source for their growth by culturing them in M9 buffer containing 0.1% benzoate. The result showed that Pseudomonas grew very well in the M9 buffer and then it was followed by Achromobacter. It means these bacteria could perfectly utilize benzoate as the carbon source for their growth. Variovorax, Sphingomonas and Cytopha could also grow in the M9 buffer (with OD600 of > 0.01 in 24 h). However, Agrobacterium, Stenotrophomonas, Spingobacterium, Herbaspirillum and Pedobacter could not grow in the M9 buffer (with OD600 of < 0.01 in 24 h), meaning that they could not utilize benzoate as the carbon source for growth (Figure 6B).
We also assigned those metagenes involved in the pathway of α-pinene degradation to a taxonomic affiliation by MEGAN. But only a small part of them (174 metagenes, 13% of total hits) in the dataset of the bacteria enriched metagenome had a definite taxonomic affiliation. The identified genera were mainly Pseudomonas, Achromobacter, Agrobacterium, Cupriavidus, Stenotrophomonas, Caulobacter, Sphingobacterium, Rhizobium, etc. (supplemental Table S2). Among them, 40 metagenes were assigned to Pseudomonas, 23 metagenes were assigned to Achromobacter and Agrobacterium. These bacteria might be the main strains for α-pinene degradation.
Xenobiotics degradation in a strain of Serratia achieved from B. xylophylus
We also downloaded the published genome sequences of Serratia sp. M24T3 from NCBI (accession: AJHJ00000000), which was achieved from B. xylophylus isolated from Pinus pinaster in Portugual43 and KEGG functional annotation was performed. Similar result was obtained from the genome, with more hits in benzoate degradation (supplemental figure S3). In addition, there are 54 genes encoding 9 enzymes involved in α-pinene degradation in the genome, only one enzyme (EC6.2.1.-) was not found in the genome of Serratia sp. (Table 2). The result indicated that bacteria associated with PWN have similar xenobiotic metabolism, even through the association types are different, such as Serratia sp. M24T3 was taken as a potential biocontrol agent for PWN43.
Discussion
Many previous works have studied bacteria associated with PWN and some bacteria were isolated and identified by culture-dependent methods24,25,26 and also by 16S rRNA gene sequences27,28,44. Comparing results obtained in this study and previously published results reveals that the composition of bacterial fauna and the abundance of each bacterium associated with each geographical PWN population were different. For example, it was reported that Pseudomonas were dominant strains in PWNs isolated in China, but Bacillus were dominant in Japan and both genera were dominant in Korea45. However, these two genera were not included in the new report of bacteria isolated from Korean PWN by Kwon and his colleagues46. In Portugal, the dominant bacteria are different among the three studies27,28,44. We think the following factors could be contribute to the different results obtained in different studies including: the material used for bacterial isolation is chips of wood (which tree species) or the nematodes (which nematode species); the culture medium (which type of medium); the isolation methods (colonies collected from the PWN trail or from the crushed PWN suspension); treatments (used or not) and its methods to exclude organisms contaminating the outer surfaces of the nematodes. Moreover, the temporal and spatial dynamics of bacterial populations could also affect the abundance of each strain. It was reported that the amount of bacteria and the predominant species changed with the symptoms procession after inoculation of PWN in the tree Pinus pinaster44 and the major bacterial population associated to the nematodes was different according to the forest area27. In our study, Stenotrophomonas was the most dominant strain in PWNs isolated in Zhejiang Province in China, which was different from previous reports25,26. Except the factors including temporal and spatial variation of bacterial community, it is very difficult to distinguish Stenotrophomonas maltophilia from the genus Pseudomonas and Xanthomonas using regular identification method, which also lead to the ambiguity. For example, S. maltophilia was initially classified as Pseudomonas maltophilia, but changed to Xanthomonas maltophilia and eventually become the type species belong to the genus Stenotrophomonas47.
As the microbiome used for this metagenomic study was isolated from nematodes of a long-cultured strain in our study, one may question that if the microbiome is bias to that in the nature environment. We compared our result with the result of a recent study, in which bacteria were obtained from strains of B. xylophilus isolated from naturally wilted pine trees in China48. We found that the composition of bacterial genera and the dominant species are similar, suggesting that the bacterial strains acquired in our study can represent the current microbiomes of PWN. Similar result was also obtained in another study, in which by comparing bacterial composition associated to PWN from symptomatic tree Pinus pinaster and from long-term preserved cultures, it was found that the main composition and the predominant genera from both source were similar28. Thus, some species remain in association with B. xylophilus, even after successive nematode generations in the laboratory, suggesting a strong specific bacteria–nematode relationship.
Even though the fauna of bacterial consortia symbiosis with PWN perhaps has a temporal and spatial variation, the ecological functions of the symbiotic bacterial consortia are similar. Comparing the two metagenomes obtained from the nematode kept in laboratory and the nematode from chips of a symptomatic tree, as well as with the bacterial genome from Portuguese PWN, the functional annotations of them were very similar. Enzymes involved in metabolism of xenobiotics degradation are abundant in the symbiotic bacteria, which formed a complex metabolic network. Benzoate (map00362) and its related degradation pathways are likely the main metabolic pathways of detoxification and each has at least one complete degradation pathway. Moreover, a complete pathway of α-pinene degradation was also observed in the bacterial metagenome (Figure 4, Table 2). Therefore, xenobiotics detoxification is an important ecological function of the bacterial consortia symbiosis with PWN.
Xenobiotics detoxification is equally important to PWN. Analysis of xenobiotic metabolisms in PWN from the genome49 and the transcriptome10 published in NCBI showed that pathways of detoxification metabolism in the nematode were obviously different from those in the symbiotic bacteria and two pathways are complementary. As previously reported, cytochrome P450 enzymes are rich in the nematode and xenobiotics degradation by cytochrome P450 (map00980, map00982) are the main pathways10,49. However, a few enzymes were involved in benzoate degradation and its related pathways. Also, only a few enzymes are involved in the pathways of limonene and pinene degradation in the genome of PWN and they could not fulfil a complete pathway of pinene degradation (Table 2). However, pinenes are the most abundant monoterpenoids in pine hosts. It was reported that crude sulphate turpentine from pine trees was mostly composed of α-pinene (60–65%) and β-pinene (25–35%)50. The secondary metabolic compounds, such as benzoic acid, benzaldehyde, phenol, phenylethanone, etc., are also relatively rich in pine trees5. Therefore, living on pine trees, the nematodes must have effective mechanisms to degrade these compounds. Based on our results, we hypothesize that PWN and its symbiotic bacteria have evolved a complete xenobiotic degradation system by a functional complementary mutualism in order to cope with various compounds produced by the secondary metabolism of the pine host.
Methods
Samples sources
Bacteria were isolated from two PWN isolates, both of which were collected from Zhejiang Province, China. One PWN isolate (ZJSS) was long-term cultured in our library and another was newly collected from chips of a pine tree that suffered from PWD in Zhejiang Province. Baermann funnel technique was employed for nematodes isolation51. Nematodes were sterilized with 5% H2O2 for 30 min to get rid of opportunistic bacteria attaching on the surface of nematodes before they were cultured for the first time. Nematodes were cultured on fungal mats of Botrytis cinerea grown on potato-dextrose agar (PDA) plates at 25°C for about ten days. Fresh cultured nematodes were collected and washed several times with double distilled water (DDW) and then used for DNA extraction and bacteria isolation.
Shotgun library construction
A shotgun genomic DNA library was constructed from genomic DNA extracted from enriched bacteria, which were isolated from the long-term cultured PWN strain in our laboratory. First, symbiotic bacteria were enriched by Nycodenz density gradient centrifugation52. Second, the enriched bacterial cells were suspended and used for DNA extraction. A typical phenol/chloroform/isoamyl alcohol method was used to extract the genomic DNA of bacteria53. DNA from triple extractions was mixed together and used for the library construction. Third, about 20 μg genomic DNA was used for a shotgun DNA library construction. DNA library preparation followed the manufacturer's instruction (Illumina), as reported previously54. Final, paired-end sequencing was performed (read length 75 bp, insert size 130 bp) using the Illumina Genome Analyzer at the Beijing Genomics Institute (Shenzhen, China).
Metagenome data analysis
The raw sequences obtained from the shotgun library were first cleaned up by trimming adapters, removing low quality reads and PCR duplication. High quality short reads were then assembled by the SOAPdenovo assembler55, with -M 3 -d 1 -u parameters. Different K values (from 21 to 63) were tested and the value 23 was selected based on the largest N50 of the assembled contigs. Contigs more than 200 bp were reserved to form a dataset, which was used for the later analysis. For functional annotation, MetaGene56 was employed to predict the ORFs of the assembled contigs. The predicted ORFs were grouped using CD-HIT-EST with the coverage over 90% and minimum identity 95% and finally the non-redundant gene set was obtained. The ORFs were translated into protein sequences using NCBI Genetic Codes11. We used BLASTp to search for the protein sequences of the predicted Genes in KEGG databases (http://www.genome.jp/kegg/), with e-value < 1e−5. The genes with KEGG annotation were assigned into KEGG pathways. For taxonomic assignment we firstly used the Illumina GA reads aligned against the known bacteria genomes (ftp.ncbi.nih.gov/genomes/Bacteria/) using SOAPaligner. Two mismatches at most were allowed in the first 35-bp region and 90% identity over the read sequence, to get the information of taxa and their reads number. Then, we used the assembled contigs to search against the dataset including all of the microbe genome sequences deposited in GenBank by BLASTn, with 1 × 10−8 and minimal 90% identity cut-off. The taxonomical level of each contig was then determined by the lowest common ancestor (LCA)-based algorithm that was implemented in MEGAN457. The taxonomic abundance was evaluated by reads number of each taxon, which was obtained by mapping all of the reads to the assembled contigs using SOAP2 with default parameters55.
To examine whether genomic DNA harvested from enriched bacteria is biased against the bacterial species that are hard to dissociate from the nematodes, we have constructed another shotgun library from the total DNA extracted from a newly isolated PWN strain directly, with no treatment of bacterial enrichment or any effort to disassociate bacteria from their host. Nematodes were first pulverised by successive freeze-thaw cycles in liquid N2 for five to six times, then the total DNA was extracted using phenol-chloroform extraction method. Library construction and sequencing was finished in the Beijing Genomics Institute (Shenzhen, China) with a Roche/454 Genome Sequencer 20 System. The clean reads were first assembled by SOAPdenovo software. Then, the assembled contigs were filtrated by eliminating the nematode sequences, with reference to the genome sequences of B. xylophilus49. The remaining assembled contigs were searched against all of the microbe genome sequences deposited in NCBI using BLASTn. Taxonomic assignment and KEGG function annotation were performed as above.
16S rRNA gene library construction and sequences analysis
The diversity of bacteria symbiosis with the PWN strain used for the metagenome was analyzed by construction of a 16S rRNA gene library30. The procedure was as following: firstly, the cultured nematodes were pulverised by successive freeze-thaw cycles in liquid N2 for five to six times and total DNA was extracted using a phenol-chloroform extraction method. Secondly, the 16S rRNA gene was amplified with primers 27F and 1492R58. To reduce the potential bias of a single experiment, DNA was extracted three times from each PWN strain and amplified with five replicas. All PCR products from the same PWN strain were mixed together for a library construction. Thirdly, the purified PCR products were ligated into the PGM-T vectors (TIANGEN, China) and then transformed into competent cells (Escherichia coli, TIANGEN, China). Lastly, transformants were plated on LB medium containing 50 μg/ml ampicillin overnight at 37°C. About 200 white colonies were picked out randomly from each transformation plate and separately grown in LB liquid medium containing ampicillin (50 μg/ml) at 37°C for 6 hours. By PCR identification, 75 positive clones containing the full-length inserts (about 1500 bp) were selected to sequence with primers T7 and SP6 using the 3730 sequencer (sequenced by SinoGenoMax Co., Ltd, China). The obtained sequences were checked for chimeras using the Chimera Check programme from the Ribosomal Database Project (RDP)(http://rdp.cme.msu.edu/). Operational Taxonomic Units (OTUs) were calculated with the software package DOTUR at a 97% similarity59. The representative sequences of each OTU were searched against the NCBI database using BLASTN to search matches.
Cultivable bacteria isolation and 16S rRNA gene identification
Nematodes were placed into 200 μl DDW and homogenised. Homogenate was cultured in Petri dishes with a LB (Luria Broth) nutrient medium and then incubated in a growth chamber at 30°C for 5 days. Then, after a serial ten-fold dilution (10−6), the cultured suspension was spread on LB medium plates. The total numbers of bacterial colonies on each plate were counted and then preliminarily categorised based on colony morphology (shape, elevation, surface, size, opacity and pigmentation). Five to ten clones of each type of isolates were picked out randomly from each plate and separately put into 400 μl LB culture fluid with a sterilized toothpick and incubated at 25°C for 24 h. Then, a 4-ml bacterial suspension of each culture was centrifuged at 12,000 rpm for 1 min and the total DNA was extracted according to a modified alkaline lysis protocol60. The 16S rRNA gene was amplified with above primers. The PCR products were purified using the EasyPure quick gel extraction kit (Transgene, China) and then directly sequenced from both ends.
Assessment of potential ability of cultivable bacteria to survive and utilize α-pinene and benzoate
To assess the potential ability of bacteria to survive the stress of α-pinene, ten identified bacterial strains were used to test. Each strain was cultured in a 1-ml liquid LB medium at 30°C for 6 hours. A 10-μl volume of each culture (with equal density) was then added to 50 ml liquid LB medium containing 0.4% α-pinene (based on the acute oral toxicity to rat, LD50 3700 mg/kg, http://www.sciencelab.com) and cultured at 30°C with 200 rpm/min. After 6 hours, 2 ml of each culture was sampled every two hours until 36 hours. The density of bacteria was assayed by ultraviolet spectrophotometer (UNICO Instruments Co., Ltd., Shanghai, China). OD600 value of each strain cultured in LB medium with no α-pinene was taken as the control treatment. Experiment repeated three times. Mean OD600 values of the three replicates were used to draw the growth curves of each bacterial strain.
Then, we tested the ability of those cultivable bacterial strains to utilize α-pinene and benzoate as the sole carbon source. A 10-μl of each above strain (with the same concentration) was added to a 50-ml M9 buffer (1 litre containing 6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, pH7.4; after autoclaving, added 2 ml 1 M MgSO4) containing 0.4% α-pinene or 0.1% benzoate (based on LD50 1714 mg/kg to rat, http://ec.europa.eu) and cultured at 25°C for 24 h (200 rpm/min). OD600 value of each culture was detected. Experimental replicates and control treatments were designed as above. OD600 value less than 0.01 indicates the bacterial strain did not grow.
Nucleotide sequence accession numbers
Metagenome data have been submitted to NCBI, accession number was SRA048563. 16S rRNA gene sequences were submitted to GenBank (accession: GU563738 - GU563762, GU569098 - GU569161). We also downloaded the genome sequences of the nematode B. xylophilus (accession: CADV01000001- CADV01010432) and the genome sequences of bacterium Serratia sp. M24T3 isolated from Portuguese B. xylophilus (accession: AJHJ00000000) from NCBI for analysis.
References
Zhao, B., Futai, K., Sutherland, J. R. & Takeuchi, Y. Pine Wilt Disease. (Springer, Kato Bunmeisha, 2008).
Mota, M. M. & Vieira, P. Pine Wilt Disease: A Worldwide Threat to Forest Ecosystems. (Springer, Netherlands, 2008).
Kong, J. O., Park, I. K., Choi, K. S., Shin, S. C. & Ahn, Y. J. Nematicidal and propagation activities of thyme red and white oil compounds toward Bursaphelenchus xylophilus (Nematoda: Parasitaphelenchidae). J. Nematol. 39, 237–242 (2007).
Oku, H. Role of phytotoxins in pine wilt disease. J. Nematol. 20, 245–251 (1988).
Takeuchi, Y., Kanzaki, N. & Futai, K. Volatile compounds in pine stands suffering from pine wilt disease; qualitative and quantitative evaluation. Nematology 8, 867–877 (2006).
Kuroda, K. Terpenoids causing tracheid-cavitation in Pinus thunbergii infested by the pine wood nematode (Bursaphelenchus xylophilus). Ann. Phytopathol. Soc. Jpn. 55, 170–178 (1989).
Kuroda, K. Mechanism of cavitation development in the pine wilt disease. Eur. J. Forest Pathol. 21, 82–89 (1991).
Zhang, H., Kanzaki, H. & Kawazu, K. Benzoic acid accumulation in the Pinus thunbergii callus inoculated with the pine wood nematode, Bursaphelenchus xylophilus. Z. Naturforsch. 52, 329–332 (1997).
Fukuda, K., Hogetsu, T. & Suzuki, K. Ethylene production during symptom development of pine-wilt disease. Eur. J. Plant Pathol. 24, 193–202 (1994).
Yan, X. et al. Comparative transcriptomics of two pathogenic pinewood nematodes yields insights into parasitic adaptation to life on pine hosts. Gene 505, 81–90 (2012).
Woyke, T. et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443, 950–955 (2006).
Ruby, E. G. Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol. 6, 752–62 (2008).
Hooper, L. V., Littman, D. R. & Macpherson, A. J. Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012).
Tremaroli, V. & Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–149 (2012).
Murfin, K. E. et al. Nematode-bacterium symbioses — cooperation and conflict revealed in the “Omics”age. Biol. Bull. 223, 85–102 (2012).
Forst, S., Dowds, B., Boemare, N. & Stackebrandt, E. Xenorhabdus and Photorhabdus Spp.: Bugs that kill bugs. Annu. Rev. Microbiol. 51, 47–72 (1997).
Goodrich-Blair, H. & Clarke, D. J. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol. Microb. 64, 260–268 (2007).
Scholl, E. H. & Bird, D. M. Computational and phylogenetic validation of nematode horizontal gene transfer. BMC Biol. 9, 9 (2011).
Bird, D. M., Opperman, C. H. & Davies, K. G. Interactions between bacteria and plant-parasitic nematodes: now and then. Intern. J. Parasit. 33, 1269–1276 (2003).
Nour, S. M. et al. Bacteria associated with cysts of the soybean cyst nematode (Heterodera Glycines). Appl. Environ. Microbiol. 69, 607–615 (2003).
Noel, G. R. & Atibalentja, N. CandidatusPaenicardinium endonii', an endosymbiont of the plant-parasitic nematode Heterodera Glycines (Nemata: Tylenchida), affiliated to the phylum Bacteroidetes. Int. J. Syst. Evol. Micr. 56, 1697–1702 (2006).
Haegeman, A. et al. An endosymbiotic bacterium in a plant-parasitic nematode: member of a new Wolbachia supergroup. Int. J. Parasitol. 39, 1045–1054 (2009).
Vandekerckhove, T. T., Willems, A., Gillis, M. & Coomans, A. Occurrence of novel Verrucomicrobial species, endosymbiotic and associated with parthenogenesis in Xiphinema americanum-Group Species (Nematoda, Longidoridae). Int. J. Syst. Evol. Microbiol. 50, 2197–2205 (2000).
Kawazu, K., Yamashita, H., Kobayashi, A. & Kanzaki, H. Isolation of pine-wilting bacteria accompanying pine wood nematode, Bursaphelenchus xylophilus and their toxic metabolites. Sci. Rep. Facul. Agr. 87, 1–8 (1998).
Zhao, B. G., Wang, H. L., Han, S. F. & Han, Z. M. Distribution and pathogenicity of bacteria species carried by Bursaphelenchus xylophilus in China. Nematology 5, 899–906 (2003).
Han, Z. M., Hong, Y. D. & Zhao, B. G. A study on pathogenicity of bacteria carried by pine wood nematodes. J. Phytopathol. 151, 683–689 (2003).
Proença, D. N. et al. Diversity of bacteria associated with Bursaphelenchus xylophilus and other nematodes isolated from Pinus pinaster trees with pine wilt disease. PLoS ONE 5, e15191 (2010).
Vicente, C. S. L., Nascimento, F., Espada, M., Mota, M. & Oliveira, S. Bacteria associated with the pinewood nematode Bursaphelenchus xylophilus collected in Portugal. Antonie van Leeuwenhoek 100, 477–481 (2011).
Zhang, Q., Tian, X., Tan, Z., Chen, G. & Xie, B. Construction and analysis of the metagenomic fosmid library for the bacteria carried by the pinewood nematode. Acta Phytopathol. Sinica 40, 381–387 (2010).
Tian, X. et al. Diversity of bacteria associated with pinewood nematode revealed by metagenome. Acta Microbiol. Sinica 50, 909–916 (2010).
Tian, X. et al. Composition of bacterial communities associated with a plant-parasitic nematode Bursaphelenchus mucronatus. Curr. Microbiol. 62, 117–125 (2011).
Zhao, B. G., Guo, D. S. & Gao, R. Observation of the site of pine wood nematodes where bacteria are carried with SEM and TEM. J. Nanjing Forest Univ. 24, 69–71 (2000).
Zhao, B. & Lin, F. Mutualistic symbiosis between Bursaphelenchus xylophilus and bacteria of the genus Pseudomonas. Forest Pathol. 35, 39–345 (2005).
Zhao, B. G., Liu, Y. & Lin, F. Effects of bacteria associated with pine wood nematode (Bursaphelenchus xylophilus) on development and egg production of the nematode. J. Phytopath. 155, 26–30 (2007).
Oku, H. Pine wilt toxin, the metabolite of a bacterium associated with a nematode. Naturwissenschaften 67, 198–199 (1980).
Tamura, H. Pathogenicity of aseptic Bursaphelenchus xylophilus and associated bacteria to pine seedlings. Jpn. J. Nematol. 13, 1–5 (1983).
Xie, L. Q. & Zhao, B. G. Post-inoculation population dynamics of Bursaphelenchus xylophilus and associated bacteria in Pine Wilt Disease on Pinus thunbergii. J. Phytopathol. 156, 385–389 (2008).
Zhao, B. G. & Li, R. G. [The role of bacteria associated with the pinewood nematode in pathogenicity and toxin–production related to pine wilt.]. Pine Wilt Disease. [Zhao B., Futai K., Sutherland J. R., & Takeuchi Y. (eds).] [250–259] (Springer, Kato Bunmeisha, 2008).
Zhu, L. et al. Pathogenicity of aseptic Bursapelenchus xylophilus. PLoS ONE 7, e38095 (2012).
Andrews, R. E., Parks, L. W. & Spence, K. D. Some effects of Douglas fir terpenes on certain microorganisms. Appl. Environ. Microbiol. 40, 301–304 (1980).
Shi, W. et al. Comparative genomic analysis of the endosymbionts of herbivorous insects reveals eco-environmental adaptations: biotechnology applications. PLoS Genet 9, e1003131 (2013).
Grzymski, J. J. et al. Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility. P. Natl. Acad. Sci. USA 105, 17516–17521 (2008).
Proença, D. N., Santo, C. E., Grass, G. & Morais, P. V. Draft genome sequence of Serratia sp. strain M24T3, isolated from pinewood disease nematode Bursaphelenchus xylophilus. J. Bacteriol. 194, 3764 (2012).
Roriz, M., Santos, C. & Vasconcelos, M. W. Population dynamics of bacteria associated with different strains of the pine wood nematode Bursaphelenchus xylophilus after inoculation in maritime pine (Pinus pinaster). Exp. Parasitol. 128, 357–364 (2011).
Kawazu, K., Zhang, H. & Kanzaki, H. Accumulation of benzoic acid in suspension cultured cells of Pinus thunbergii Parl. in response to phenylacetic acid administration. Biosci. Biotech. Bioch. 60, 1410–1412 (1996).
Kwon, H. R. et al. Suppression of pine wilt disease by an antibacterial agent, oxolinic acid. Pest Manag. Sci. 66, 634–639 (2010).
Palleroni, N. & Bradbury, J. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 198. Int. J. Syst. Bacteriol. 43, 606–609 (1993).
Wu, X.-Q. et al. Specific and functional diversity of endophytic bacteria from pine wood nematode Bursaphelenchus xylophilus with different virulence. Int. J. Biol. Sci. 9, 34–44 (2013).
Kikuchi, T. et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7, e1002219 (2011).
FRIDGE (Fund for Research into Industrial Development, Growth and Equity). Study into the establishment of an aroma and fragrance fine chemicals value chain in South Africa, Part Three: aroma chemicals derived from petrochemical feedstocks. NEDLAC (National Economic Development and Labor Council). 22–63 (2004).
Viglierchio, D. R. & Schmitt, R. V. On the methodology of nematode extraction from field samples: Baermann funnel modifications. J. Nematol. 15, 438–444 (1983).
Ikeda, S. et al. Development of a bacterial cell enrichment method and its application to the community analysis in soybean stems. Microbiol. Ecol. 58, 703–714 (2009).
Moore, D. D. & Dowhan, D. Preparation and analysis of DNA. Current Protocols in Molecular Biology 2.0.1–2.0.3 (2002).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Li, R. et al. De novo assembly of the human genomes with massively parallel short read sequencing. Genome Res. 10.1101/gr.097261.109 (2009).
Noguchi, H., Park, J. & Takagi, T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Research 34, 5623–5630 (2006).
Huson, D. H., Mitra, S., Ruscheweyh, H.-J., Weber, N. & Schuster, S. C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21, 1552–1560 (2011).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 679–703 (1991).
Schloss, P. D. & Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microb. 71, 1501–1506 (2005).
Feliciello, I. & Chinali, G. A modifid alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia coli. Anal. Biochem. 12, 394–401 (1993).
Acknowledgements
We are very grateful to Dr. Li for the gifts of Canadian and Portuguese nematodes. This work was supported by the National Key Basic Research and Development Program of China (grant no.2009CB119200) and a Discovery Grant from The Natural Sciences and Engineering Research Council of Canada (NSERC) to NC.
Author information
Authors and Affiliations
Contributions
B.Y.X. and X.Y.C. conceived and designed the experiments; X.L.T. and Z.C.M. performed the experiments; Y.S.W., R.M.L., X.L.T. and X.Y.C. analyzed the data; X.Y.C. and X.L.T. wrote the paper; N.C. and B.Y.X. edited and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
dataset 1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Cheng, XY., Tian, XL., Wang, YS. et al. Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci Rep 3, 1869 (2013). https://doi.org/10.1038/srep01869
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01869
This article is cited by
-
Thauera sp. for efficient nitrate removal in continuous denitrifying moving bed biofilm reactor
Bioprocess and Biosystems Engineering (2024)
-
The silkworm (Bombyx mori) gut microbiota is involved in metabolic detoxification by glucosylation of plant toxins
Communications Biology (2023)
-
Bioprospection of the bacterial β-myrcene-biotransforming trait in the rhizosphere
Applied Microbiology and Biotechnology (2023)
-
The microbial community associated with Parascaris spp. infecting juvenile horses
Parasites & Vectors (2022)
-
An integrated host-microbiome response to atrazine exposure mediates toxicity in Drosophila
Communications Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.