Abstract
Bioactive compounds are secondary metabolites of plants. They offer diverse pharmacological properties. Peganum harmala is reported to have pharmaceutical effects like insecticidal, antitumor, curing malaria, anti-spasmodic, vasorelaxant, antihistaminic effect. Rosa brunonii has medicinal importance in its flower and fruits effective against different diseases and juice of leaf is reported to be applied externally to cure wounds and cuts. Dryopteris ramosa aqueous leaf extract is used to treat stomach ulcers and stomachaches. Each of these three medicinal plants have been indicated to have anticancer, antiviral, antioxidant, cytotoxic and antifungal effects but efficacy of their bioactive compounds remained unexplored. Study was aimed to explore In-vitro and In-silico anticancer, antiviral, antioxidant, cytotoxic and antifungal effects of bioactive compounds of above three medicinal plants. DPPH and ABTS assay were applied for assessment of antioxidant properties of compounds. Antibacterial properties of compounds were checked by agar well diffusion method. Brine shrimp lethality assay was performed to check cytotoxic effect of compounds. Molecular docking was conducted to investigate the binding efficacy between isolated compounds and targeted proteins. The compound isomangiferrin and tiliroside presented strong antioxidant potential 78.32% (± 0.213) and 77.77% (± 0.211) respectively in DPPH assay while harmaline showed 80.71% (± 0.072) at 200 µg/mL in ABTS assay. The compound harmine, harmaline and PH-HM 17 exhibited highest zone of inhibition 22 mm, 23 mm, 22 mm respectively against Xanthomonas while Irriflophenone-3-C-β- D-glucopyranoside showed maximum zone of inhibition 34 mm against E. coli. The compound isomangiferrin and vasicine contained strong antibacterial activity 32 mm and 22 mm respectively against S. aureus. The compound mangiferrin, astragalin, tiliroside, quercitin-3-O-rhamnoside showed maximum inhibitory zone 32 mm, 26 mm, 24 mm and 22 mm respectively against Klebsiella pneumoniae. Highest cytotoxic effect was observed by compound tiliroside i.e. 95% with LD50 value 73.59 µg/mL. The compound tiliroside showed the best binding mode of interaction to all targeted proteins presenting maximum hydrophobic interactions and hydrogen bonds. The binding affinity of tiliroside was − 17.9, − 14.9, − 14.6, − 13.8, − 12.8 against different proteins 6VAR, 5C5S, IEA3, 2XV7 and 6LUS respectively. Bioactive compounds are significant natural antioxidants, which could help to prevent the progression of various diseases caused by free radicals. Based on molecular docking we have concluded that phytochemicals can have better anticancer and antiviral potential.
Similar content being viewed by others
Introduction
Bioactive compounds are secondary metabolites of plants. They offer diverse pharmacological properties due to great variety in their structures and play an important role in prevention and treatment of wide array of diseases from fever to dreadful cancers1. Peganum harmala L. is commonly known as Syrian rue and Harmal, belongs to family Zygophyllaceae2. P. harmala have pharmaceutical effects like insecticidal, antitumor, antimalarial, anti-spasmodic, vasorelaxant, antihistaminic effect. It shows different activities such as antioxidant, cytotoxic, antimutagenic, antifungal, herbicidal and fungicidal3,4,5. Plants also have anti-parasitic activity, anti-tumor, and hallucinogenic activities6,7. Fruit has been used for treatment of different diseases like asthma, jaundice, and colic8. Different bioactive compounds harmine, harmol, harmaline, and harmalol are beta-carboline alkaloids that are typically found in roots and seeds9. The isoxazole derivatives of harmine show anti-alzheimer activity10. Roots and seed extract of P. harmala were most biologically active, having strong anticancer and cytotoxic effects, presumably due to the presence of phytochemicals such flavonoids and phenolics11.
Rosa brunonii L. is a member of the Rosaceae family and is known as the 'Himalayan musk rose', found in Himalayan region and Kashmir12. It is commonly known as ‘jungli gulap’ or ‘tarni’13. Flower and fruits of R. brunonii have medicinal importance as well as used as food14. Flower and fruits of this plant are effective against different diseases and digestive problems15. Crude extract from flower of R. brunonii show strong anti-oxidant potential. Leaf juice has been applied externally to cure wounds and cuts12,16. Different compounds extracted from fruit of R. brunonii like astragalin, quercetin-3-O-rhamnoside and tiliroside show strong anti-oxidant properties15. The phytochemicals of R. brunonii have cytotoxic effects17. The methanolic crude extracts of R. brunonii have cytotoxic effect and its fruits extract have antibacterial effect18.
Dryopteris ramosa is commonly known as Pakha, the member of family Dryopteridacea. It thrives in wet, shady environments. The fronds of D. ramosa are edible and can be taken orally for antibacterial and aphrodisiac effects19. The phytochemicals isolated from fronds of Dryopteris ramosa have strong anti-microbial effect20. Mangiferin and isomangiferin have been shown to exhibit antibacterial, immunomodulatory, anti-diabetic, anti-oxidative, anthelmintic, anti-inflammatory, anti-HIV, anticancer, anti-viral, anti-allergic, and macrophage-inducing actions21. The compound mangiferrin has hepatoprotective and analgesic abilities22. The isomangiferrin and mangiferrin have strong antioxidant potential18. Iriflophenone-3-C-β-d Glucopyranoside showed strong antibacterial potential against E. coli K. pneumoniae, S. aureus and low cytotoxic activity23. The present study aimed to investigate purified compounds for their antioxidant, antibacterial, cytotoxic and In silico analysis of purified compounds of medicinal plants. The study is novel in its type regarding combination studies of these compounds by in vitro and in silico assays for elucidating the pharmacological potential of native species of Pakistan against dreadful diseases caused by pathogenic bacteria as well as free radicals. This study is of particular importance in Pakistan, where the local population predominantly depends on these medicinal plants for the treatment of various diseases.
Results and discussion
New therapeutic compounds can be synthesized using secondary metabolites found in medicinal plants. The types of secondary metabolites in solvent extract may vary depending on the solvent and extraction method. Different compounds like harmine, harmaline, vasicine, vasicinone (P. harmala) mangiferrin, isomangiferrin, irriflophenone -3-C-β-D D-glucopyranoside (D. ramosa) tiliroside, astragalin and quercitin-3-O rahmnoside (R. brunonii) were subjected for evaluation of their antioxidant, cytotoxic and antimicrobial potential. Molecular docking of these compounds was also done against different diseases (cancer, hepatitis B virus, influenza virus and SARS Covid 19).
DPPH and ABTS assays were used to check antioxidant potential. In DPPH assay all compounds showed significant free radical scavenging potential but the compound isomangiferrin and tiliroside exhibited highest antioxidant potential of 78.32% (± 0.213) and 77.77% (± 0.211) at 200 µg/mL (Fig. 1). In term of IC50 values isomangiferrin showed IC50 value of 93.85 µg/mL and tiliroside showed IC50 value 78.77 µg/mL that were less than ascorbic acid showed IC50 value 97.065 µg/mL. The low IC50 means strongest scavenging potential. It means that scavenging capacity of these compounds were strongest than ascorbic acid. Quercitin-3-O rhamnoside exhibited lowest 50% free radical scavenging potential with IC50 value 185.27 µg/mL concentration (Table 1). The IC50 value of compounds was calculated by linear regression equation (Table 1). The dark violet color changed to light yellow also predicted antioxidant potential of compounds. Our study is in correspondence to a previously conducted study in which isomangiferrin and tiliroside showed strongest antioxidant potential in DPPH assay as compared to other compounds15,21,24,25.
Another assay 2,2 -azino-bis-3-ethyibenzothiazoline-6-sulfonic acid was used to determine the free radical scavenging potential of compounds. All compounds showed significant free radical scavenging potential by ABTS assay. The maximum activity was shown by compound of P. harmala (harmaline) i.e. 80.71% with IC50 value 15.79 µg/mL. In terms of percentage and IC50 value others compounds (Astragalin, mangiferrin, Irriflophenone-3-C-β- D-glucopyranoside, tiliroside, vasicine, Quercetin-3-O- rahmnoside, isomangiferrin and harmine) showed significant free radical scavenging potential i.e. 78% (49.27 µg/mL), 73% (110.33 µg/mL), 71% (112.63 µg/mL), 70% (101.64 µg/mL), 69% (114.82 µg/mL), 67% (111.60 µg/mL), 62% (124.19 µg/mL) and 61.33% (148.26 µg/mL) respectively at 200 µg/mL (Fig. 2). More than 60% antioxidant activity was shown by all compounds. Ascorbic acid was used as standard and showed highest percentage (90%) with lowest IC50 value (14.77 µg/mL) as compared to all compounds (Fig. 2). The IC50 value of harmaline was near to IC50 value of ascorbic acid. The color was changed from bluish green to colorless that indicated the antioxidant activity of compounds. The antioxidant potential of all compounds and standard (Ascorbic acid) was expressed in terms of IC50 value. The linear regression equation was used to calculate IC50 values (Table 2).There was difference in antioxidant potential of compounds both in ABTS and DPPH assay may be due to presence to different radicals and solvent. The percentage scavenging activity increased with increase in concentration26,27,28.
Different compounds showed different inhibitory zones against different bacterial strains. Previously pharmacological actions of P. harmala including anti-oxidant, anti-tumor, hypoglycemic effect, leukemia healing, anti-inflammatory and analgesic properties, cytotoxic anti-tumor, and anti-microbial effects has been reported by Asgarpanah and Ramezanloo29. In present study the compound harmine and harmaline exhibited highest zone of inhibition 22 mm and 23 mm respectively against Xanthomonas while vasicine showed 22 mm against S. aureus. Irriflophenone-3-C-β- D-glucopyranoside showed maximum zone of inhibition 34 mm, 31 mm, 30 mm and 28 mm against E. coli, Salmonella, K. pneumonia and S. aureus respectively while less active against P. aeruginosa, S. epidermitis and Xanthomonas with inhibitory zones 8 mm,16 mm and 19 mm respectively. Mangiferrin was more active against K. pneumonia (32 mm) and Salmonella (28 mm) and weak against P. aeruginosa (10 mm), E. coli (15 mm). The compound isomangiferrin contained strong antibacterial activity against S. aureus (32 mm) and K. pneumonia (28 mm) while lowest against 5 mm against P. aeruginosa. Astragalin, tiliroside and quercetin-3-O rhamnoside) showed strongest activity against K. pneumonia with inhibitory zone i.e. 26 mm, 24 mm, and 22 mm respectively. Similarly strongest antibacterial activity of Irriflophenone-3-C-β-D-glucopyranoside, mangiferrin and isomangiferrin was observed21,23. The compound of R. brunonii i.e. astragalin, tiliroside, quercitin-3-O-rhamnoside showed maximum inhibitory zone 26 mm, 24 mm, and 22 mm respectively against Klebsiella pneumoniae, while lowest against P. aeruginosa. Comparison of compounds antibacterial potential against different strains (Fig. 3). In previous study it has been observed that methanolic fruit's extract of R. Brunonii showed highest antibacterial potential against Klebsiella pneumoniae and lowest against Bacillus subtilis18.
For cytotoxic assessment of compounds brine shrimp lethality assay was used. The compound tiliroside exhibited 95% mortality rate with LD50 value 73.59 µg/mL highest mortality rate as compared to all other compounds. Harmaline showed 80% cytotoxic effect while harmine and Irriflophenone-3-C-β-D-glucopyranoside showed 65% mortality at highest 200 µg/mL concentration. The 55% was observed by vasicine, isomangiferrin and astragalin at 200 µg/mL. The lowest 50% cytotoxicity was showed by compound mangiferrin whereas 60% cytotoxic effect was observed by quercetin-3-O rhamnoside at 200 µg/mL (Fig. 4). Nicotine was used as standard. It showed 100 percent mortality rate at 200 µg/mL with lowest LD50 value 30.69 µg/mL among all compounds (Table 3). In previous literature cytotoxicity of mangiferrin and isomangiferrin has been observed21. Antibacterial, antioxidant and cytotoxic effects of some compounds were checked for the first time, but in previous literature same work has been done on whole plants and different fractions of these plants (P. harmala, D. ramosa and R. brunonii). These above different compounds were isolated from these plants. That’s why compounds also exhibited such potentials like antibacterial, antioxidant and cytotoxic potentials.
In the process of drug discovery, computational innovations had a substantial impact. Computational biology emerged as an interdisciplinary approach integrating the computational techniques with biological systems to target different disease related challenges30.Virtual screening methods are frequently used to reduce the cost and time of medication development. Virtual screening predicts both binding modes and affinities of receptor and ligand based on docking and scoring of each molecule in a data set. Molecular docking has aided important drug discovery procedures. Docking approaches aid in the identification of correct ligand poses in a protein's binding pocket as well as the prediction of ligand–protein affinity. Finally, docking is a three-dimensional process for fitting two molecules together31,32,33,34,35.
In this study, molecular docking of different compounds that were previously reported from three medicinal plants was also done with target proteins (anticancer and antiviral). Similar study was reported by Gayathiri et al.,30 in Sauropus androgynus (L.) Merr against inflammation and cancer. The biochemical compounds were taken ligand to evaluate the antiviral and anticancer efficacy of these compounds. The 3D structures of antiviral and anticancer proteins were obtained from RCSB Protein Data Bank. Docking results were represented in the form of e-negative values (Tables 4 and 5). The arrangements of ligands (compounds) were based on ligand–protein binding energies. In molecular docking, higher e-negative values represent high binding affinity between the receptor and ligands molecules that indicated the higher efficacy of bioactive compound.
In current study the tiliroside compound that was extracted from plant Rosa brunonii showed best affinity towards all proteins (anti-cancer and anti-viral). In case of anticancer protein 5C5S tiliroside showed binding energies of − 14.9 kcal/mol and − 13.8 kcal/mol in 2XV7 anticancer protein (Table 4). Tiliroside showed binding affinities of − 14.6, − 17.9 and − 12.8 kcal/mol in case of influenza virus protein 1EA3, hepatitis virus protein 6VAR and SARS Covid- 19 protein 6LUS respectively (Table 5). The lowest binding affinity was shown by the compounds of Peganum harmala. Vasicine showed binding energies − 6.5, − 7.0, − 7.5, − 7.6 kcal/mol in case of SARS Covid- 19 protein 6LUS, anticancer protein (2XV7), anticancer protein (5C5S) and hepatitis virus protein (6VAR) respectively. In case of influenza virus protein (1EA3), compound harmaline showed lowest binding affinity of − 7.2 kcal/mol.
The sequence of compounds according to binding energy in case of anticancer protein (2XV7) was tiliroside > Irriflophenone-3-C-β- D-Glucopyranoside > mangiferrin > quercitin-3-O-rhamnoside > astragalin > isomangiferrin > harmine > harmaline > vasicine (Table 4). In case of second anticancer protein (5C5S) the sequence of compounds according to their binding energy was tiliroside > Irriflophenone-3-C-β- D-Glucopyranoside > astragalin > mangiferrin > isomangiferrin > quercitin-3-o-rhamnoside > harmine > harmaline > vasicine as shown in Table 4. The compounds sequence according to binding affinities in hepatitis protein (6VAR) case was Tiliroside > mangiferrin > quercitin-3-o-rhamnoside > astragalin > Irriflophenone-3-C-β- D-Glucopyranoside > Isomangiferin > harmine > harmaline > vasicine, while in case of influenza virus protein (1EA3) the sequence followed was Tiliroside > mangiferrin > Irriflophenone-3-C-β- D-Glucopyranoside > quercitin-3-o-rhamnoside > Isomangiferin > astragalin > vasicine > harmine > harmaline shown in Table 5.
For the first time, molecular docking of these compounds was carried out, targeting anticancer and antiviral proteins. This study is of particular importance in Pakistan, where the local population predominantly depends on these medicinal plants for the treatment of various diseases. Our research is a crucial step in drug development, especially in the context of SARS-CoV-2, a highly contagious disease prevalent today. Herbal medicines are not only cost-effective but also demonstrate long-term effectiveness with fewer side effects compared to chemically synthesized medicines. Additionally, previous studies have demonstrated the effectiveness of these plants in vitro in a variety of ways, including anticancer, antioxidant, antibacterial, and antiviral properties. The binding energies obtained by docking of 6LUS protein with ligands (compound) in a sequence (1) tiliroside (2) quercitin-3-o-rhamnoside (3) Astragalin (4) isomangiferrin (5) mangiferrin (6) Irriflophenone-3-C-β- D-Glucopyranoside (7) harmaline (8) harmine (9) vasicine was − 12, − 12.6, − 12.0, − 11.6, − 11.2, − 11.1, − 6.7, − 6.6, − 6.5 (Table 5). First time molecular docking of these compounds was done but in previous study, in vitro analysis of these plants on such activities like anticancer, antioxidant, antibacterial and antiviral has been exhibited2,11,15,18,21,29,36,37,38,39.
Conclusion
Plant derived bioactive compounds have several therapeutic effects. Present study is highlighting the importance of bioactive compounds of P. harmala, D. ramosa and R. brunonii with significant antibacterial, cytotoxic and antioxidant potential in vitro and in silico that justified the ethnomedicinal importance of these plants. The compound tiliroside (R. brunonii) showed highest (95%) cytotoxic effect on brine nauplii among all compounds. The current study also explored the interaction of bioactive compounds to inhibit different cancer (2XV7 & 5C5S) and viral (Hepatitis 6VAR, SARS Covid 19 6LUS & Influenza IEA3) proteins. The results from this study displayed that compound tiliroside demonstrated the high binding affinity towards all cancer and viral proteins as compared to all other compounds. These bioactive compounds are significant sources of natural antioxidants, which could help to prevent the progression of various diseases, caused by free radicals i.e. certain cancers. In conclusion these results suggest that bioactive compounds may be helpful for the development of newer and more effective pharmaceutical components which may have lesser adverse effects.
Material and methods
Isolated and purified compounds from different medicinal plants were used to test biological activities. The compounds were Harmine, Harmaline, vasicinone (PH 17) and Vasicine from P. harmala, Tiliroside, Astragalin, Quercetin-3-O-rhamnoside from R. brunonii and Mangiferin, Isomangiferin Irriflophenone-3-C-β-D-glucopyranoside from D. ramosa. These compounds were provided by Phytochemical laboratory of PMAS-Arid Agriculture University Rawalpindi, Pakistan.
Antioxidant assessment
The antioxidant activities of compounds were analyzed by DPPH Free Radical Scavenging Assay and ABTS Assay.
DPPH (2–2-diphenyl-1-picrylhydrazyl) Free Radical Scavenging Assay
The DPPH assay was performed according to the protocol of Ghasemi et al.40 with little modifications. For DPPH solution, 3 mg of DPPH powder was dissolved in 100 mL methanol. 4 mg of compounds were dissolved in 5 mL methanol to obtained stock solution of compounds. Different serial dilutions (25, 50, 100, and 200 μg/mL) were prepared. The 3 mL DPPH methanolic solution was added to 400 μL compounds methanolic solution. The mixture was continuously shaken and stored at room temperature under shade for exactly 30 min. The absorbance of reaction mixture was measured at 515 nm spectrophotometrically. The dark violet color changed to light yellow color. The experiment was repeated three times and percentage inhibition was calculated as follows.
IC50 value was calculated by linear regression equation that was obtained by plotting concentrations against percentage scavenging activity.
ABTS Assay (2,2 -azino-bis-3-ethyibenzothiazoline-6-sulfonic acid)
The ABTS test was performed using technique with some alternations41. The stock solutions (7 mM ABTS solution in 100 mL distilled water) and (2.4 mM potassium persulfate in 100 mL distilled water) were prepared. The ABTS solution was prepared by mixing equal parts of the two standard stock solutions and left them to react for 14 h at 25 °C in the dark. For compound solution, 4 mg of compounds were dissolved in 5 mL distilled water. Different concentrations (25, 50, 100 & 200 μg/mL) were prepared by using this stock solution. 400 μL compounds solution was allowed to react with 3 mL of the ABTS solution and the absorbance was taken after 30 min. Ascorbic acid was used as positive control. Color was changed from bluish green to colorless. The percentage scavenging potential was calculated by using following formula.
Absorbance blank is absorbance of control (without compound) and Absorbance sample is absorbance of sample (compound).
IC50 value was calculated by linear regression equation that was obtained by plotting concentrations against percentage scavenging activity.
Antibacterial activity
Antibacterial activity of compounds was determined by agar well diffusion method42. Antibacterial activity of different compounds was tested against seven clinical pathogens (Klebsiella pneumonia, Escherichia coli, Xanthomonas, S. aureus, Salmonella, P. aeruginosa and S. epidermitis). The nutrient agar medium was prepared by dissolving 6 g agar nutrient powder in 250 mL distilled water. The media was autoclaved and poured into pre-labeled petri plates under sterile condition. Then rinsed these plates with bacterial suspension before solidification. Allowed it to solidify for 15 min. Compound solution of 0.6 mg/mL was used. Cefixime with 1 mg/1 mL was used as positive control whereas DMSO was used as negative control. After solidification made wells of 6 mm using sterile cork borer. Each well was sealed with 20 μL of agar. After sealing 40 μL of each compound solution was added in well. The petri plates were sealed by para-film and kept in incubator for 24 h at 37 °C. The experiment was run in triplicates. The zone of inhibition of bacterial growth was measured with scale in mm.
Cytotoxicity assessment
Brine shrimp lethality assay (BSLA) was used to check out the cytotoxic potential of different compounds of plant species by using the protocol described by Ishaque et al.18 with several modifications. Artificial sea water was prepared by dissolving 38 g sea salt in 1000 mL distilled water. This sea water was poured in a two-chamber container. One chamber was big and other was small. Added brine shrimp’s eggs on small side of chamber. Small side of chamber was covered and on other side provided illumination with electric lamp. Electric lamp was used to attract brine shrimp’s larva photo-tactically. Kept this for 24 h at room temperature for hatching of eggs. After 24 h brine shrimp’s larva matured to nauplii. Picked these nauplii from illuminated side with glass capillary tube for further process. The compounds were dissolved in Dimethyl sulfoxide (DMSO) and different concentrations of these compounds were prepared (25, 50, 100 and 200 μg/mL). Nicotine was used as positive control and same concentrations were made. Artificial sea water was used as a negative control. These concentrations were poured in vials and final volume of 5 mL was made by adding sea water. Added phototropic 20 nauplii in each vial with glass capillary tube. Then these vials were kept at room temperature for 24 h for incubation. After that alive nauplii were calculated. Mortality rate of brine shrimps was measured by using formula:
LC50 was calculated by drawing graph between percentage mortality and concentrations using excel.
In silico analysis
Plant extracts possess thousands of bioactive chemicals, and determining the biological efficacy of each one in laboratory is too time consuming43. Therefore in silico evaluation of bioactive compounds from medicinal plants targeting cancerous and viral proteins has been done. Following steps were involved in Insilico analysis.
Molecular docking studies
The "key and lock" theory is utilized in molecular docking to discover the best match orientation for ligand and protein44. The structures of compounds were obtained from Pub Chem in SDF format. The phytochemicals show potential as drug candidates against the selected proteins by satisfying the Lipinski rule. Which entails a molecular weight below 500 g/mol, fewer than 10 H-bond acceptors, and fewer than 5 H-bond donors. Molecular weight, indicative of density, size, volume, and mass, is a key factor for therapeutic agents45. Furthermore, adherence to other rules, such as a molar refractivity between 40 and 130 and having fewer than 10 rotatable bonds, additional supports the drug-likeness46. Lipinski's rule-of-five, which connects physicochemical and pharmacokinetic indices, underlines that an orally active drug-like compound should not defy more than one of the subsequent criteria: hydrogen bond donors not exceeding 5, hydrogen bond acceptors not exceeding 10, molecular weight not exceeding 500 Da, and octanol–water partition coefficient (log P) not exceeding 5. Importantly, all selected phytochemicals adhere to these criteria. Anticancer (2XV7 and 5C5S) and antiviral proteins (hepatitis 6VAR, influenza 1EA3, SARS Covid-19 6LUS) were obtained from protein database. The target protein and ligands were synthesized according to normal ligand and protein preparation procedures, and the protein and ligand files were sent to Auto Dock vina. Each ligand's binding energy and binding contacts were determined. The docked complexes were studied using Ligplus.
Ligand preparation
The 3D structures of compounds (ligands) were obtained from Pub Chem compound database at NCBI in SDF format (Table 6). Then changed these SDF format structures to PDB format using pymol. Ligands were prepared by removing water molecules, adding hydrogen bonds, and required charges (Kollman and Gasteiger) using AutodockTools-1.5.7. Then saved structures of compounds in PDBQT format and used for docking by using AutodockTools-1.5.747.
Protein preparation
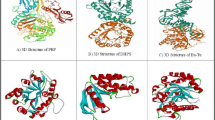
The crystal structures of different proteins were retrieved from RCSB Protein Data Bank in PDB format48. These proteins were anticancer (2XV7 & 5C5S) and antiviral (influenza 1EA3 & hepatitis 6VAR) (Fig. 5). These raw PDB proteins have only water molecules, metal ions, heavy atoms, and co-factors. These protein structures contained no information about formal atomic charges, bond orders and topologies so could not be used for molecular docking studies. So removed all water molecules, added polar hydrogen atoms and charges (Gasteiger and Kollman) in protein crystal structure using AutodockTools-1.5.7. Then optimized structure by assigning bond angles, bond orders and topology. Saved these structures in PDBQT format for further analysis.
The generation of grid box
Pre-calculated grid map is important in Auto Dock. Grid map helps in calculation of docking score. The docking evaluation results were analyzed by determining the amino acids in the active site. A three -dimensional grid box was created at maximum on proteins, to allow ligands to be docked on all areas of the receptor49. To generate grid box, selected both proteins receptor (5c5s.pdbqt, 6var.pdbqt, 2xv7.pdbqt, 1ea3 & 6lus.pdbqt) and saved ligands (compounds) in pdbqt format. Grid map was generated in such a way that both ligand and receptor fit into this three-dimensional matrix. Adjusted grid box size for X,Y,Z dimensions. The space was adjusted as 0.5 A that was nearly quarter of length for carbon single bonding atom. In the center of grid box ligand was present and was well-adjusted inside the active side of receptor proteins. Then exported the grid box dimension as grid.txt format. Then saved it in respective file for further processing.
Use of command prompt
Command Prompt is command line interpreter tool. The command is issued at the command prompt to examine the ligand-receptor binding affinity. The command prompt was open on desktop. Then provide command as “cd space the open folder of compound and add the link (E:\important\Molecular Docking\complete\cancer 2XV7) then enter “ and go to computer, open local disk C, open programme file, then open “The Scripps Research Institute” open vina, copy link and added this link (C:\Program Files\The Scripps Research Institute\Vina). After that insert \, click on double tab, space, –config, space, grid.txt, space, –log, space, log.txt and press the enter button. After that docking results of compounds were obtained after analyzing of bonding score.
The analysis of protein–ligand complex by Ligplus
For assessing and characterization of interactions in our docked protein–ligand complex, LIGPLOT + version v.2.2.4 was used to visualize compounds with the best binding affinities. The hydrophobic and polar interactions between both the ligand and the target protein were visualized using this software (Figs. 6, 7, 8, 9, 10). The ligand.pdbqt and protein.pdbqt opened in pymol at same time. Then clicked on file and exported molecule, given name as complex, saved in pdb format in respective folder. Again clicked on file, exported image as PNG, saved and give any name. The image.oxps was converted online to jpg in respective folder. This method was used to show and generate interactions between compounds.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DPPH:
-
2-2-Diphenyl-1-picrylhydrazyl
- ABTS:
-
2,2-Azino-bis-(3-ethylbenzothiazoline-6-sulphonic acid)
- BSLA:
-
Brine shrimp lethality assay
- LD50 :
-
Median lethal dose.
- IC50 :
-
Half maximal inhibitory concentration
- (DMSO):
-
Dimethyl sulfoxide
- PM17:
-
Vasicinone
References
Azmir, J. et al. Techniques for extraction of bioactive compounds from plant materials: A review. J. Food Eng. 117(4), 426–436 (2013).
Benzekri, R. et al. Anti HSV-2 activity of Peganum harmala (L.) and isolation of the active compound. Microbial Pathogenesis 114(3), 291–298 (2018).
Goel, N., Rao, H., Durmer, J. S. & Dinges, D. F. Neurocognitive consequences of sleep deprivation. Semin. Neurol. 29(4), 320 (2009).
Bemis, D. L., Capodice, J. L., Gorroochurn, P., Katz, A. E. & Buttyan, R. Anti-prostate cancer activity of a β-carboline alkaloid enriched extract from Rauwolfia vomitoria. Int. J. Oncol. 29(5), 1065–1073 (2006).
Shrivastava, S., & Dash, D. Applying nanotechnology to human health: Revolution in biomedical sciences. J. Nanotechnol., 2009 (2009)
Cao, R. et al. DNA binding properties of 9-substituted harmine derivatives. Biochem. Biophys. Res. Commun. 338(3), 1557–1563 (2005).
Ayoob, I. et al. Phytochemical and cytotoxic evaluation of peganum harmala: structure activity relationship studies of harmine. ChemistrySelect 2(10), 2965–2968 (2017).
Bukhari, A. A. Investigation of the electro-coagulation treatment process for the removal of total suspended solids and turbidity from municipal wastewater. Biores. Technol. 99(5), 914–921 (2008).
Herraiz, T., González, D., Ancín-Azpilicueta, C., Arán, V. J. & Guillén, H. β-Carboline alkaloids in Peganum harmala and inhibition of human monoamine oxidase (MAO). Food Chem. Toxicol. 48(3), 839–845 (2010).
Rah, B. et al. Design and synthesis of antitumor heck-coupled Sclareol analogues: modulation of BH3 family members by SS-12 in autophagy and apoptotic cell death. J. Med. Chem. 58(8), 3432–3444 (2015).
Sadaf, H. M. et al. Analysis of Peganum harmala, Melia azedarach and Morus alba extracts against six lethal human cancer cells and oxidative stress along with chemical characterization through advance Fourier Transform and Nuclear Magnetic Resonance spectroscopic methods towards green chemotherapeutic agents. Saudi Pharmaceutical J. 29(6), 552–565 (2021).
Abbasi, A. S., Rehman, K. U. & Bibi, A. Islamic management model. Afr. J. Bus. Manag. 4(9), 1873–1882 (2010).
Abbasi, A. M., Khan, M. A., Ahmad, M. & Zafar, M. Springer science & business media. Med. Plant Biodivers. Lesser Himalayas-Pakistan 5(7), 1545–1580 (2011).
Khan, M. P. Z. et al. Ethnomedicinal uses of edible wild fruits (EWFs) in Swat Valley, Northern Pakistan. J. Ethnopharmacol. 173(5), 191–203 (2015).
Ishaque, M., BiBi, Y., Valant-Vetschera, K. M., Schinnerl, J. & Bacher, M. Fruits of Rosa Brunonii–a source of antioxidant phenolic compounds. Nat. Product Commun. 12(11), 567–590 (2017).
Amjad, M. S. et al. Descriptive study of plant resources in the context of the ethnomedicinal relevance of indigenous flora: A case study from Toli Peer National Park, Azad Jammu and Kashmir, Pakistan. PloS One 12(2), e0171896 (2017).
Ahmad, E. et al. Rosa brunonii Lindely fruit as a new protective agent evaluated against Rif/INH induced toxicity in rats. Pak. J. Pharm. Sci. 33(2 Suppl), 805–814 (2020).
Ishaque, M. et al. Xanthone C-glycosides isomers purified from Dryopteris ramosa (Hope) C. Chr. with bactericidal and cytotoxic prospects. Saudi J. Biol. Sci. 29(2), 1191–1196 (2022).
Ahmad, K. S. & Habib, S. Indigenous knowledge of some medicinal plants of Himalaya region, Dawarian village, Neelum valley, Azad Jammu and Kashmir, Pakistan. Univ. J. Plant Sci. 2(2), 40–47 (2014).
Alam, F., Khan, S. H. A. & Asad, M. H. H. B. Phytochemical, antimicrobial, antioxidant and enzyme inhibitory potential of medicinal plant Dryopteris ramosa (Hope) C. Chr. BMC Complementary Med. Therapies 21(1), 1–10 (2021).
Ishaque, M., Bibi, Y., Qayyum, A. & Iriti, M. Isolation and structural confirmation of xanthone isomers from Dryopteris ramosa (Hope) C. Chr. and their in vitro antioxidant mechanism. Arab. J. Sci. Eng. 46(6), 53 (2021).
Sellamuthu, P. S., Arulselvan, P., Muniappan, B. P., Fakurazi, S. & Kandasamy, M. Mangiferin from Salacia chinensis prevents oxidative stress and protects pancreatic β-cells in streptozotocin-induced diabetic rats. J. Med. Food 16(8), 719–727 (2013).
Ishaque, M. et al. Iriflophenone-3-C-β-d Glucopyranoside from Dryopteris ramosa (Hope) C. Chr. with promising future as natural antibiotic for gastrointestinal tract infections. Antibiotics 10(9), 1128 (2021).
Luo, W. et al. Update: Innate lymphoid cells in inflammatory bowel disease. Dig. Dis. Sci. 67(1), 56–66. https://doi.org/10.1007/s10620-021-06831-8 (2022).
Yan, J. et al. Rewiring chaperone-mediated autophagy in cancer by a prion-like chemical inducer of proximity to counteract adaptive immune resistance. Drug Res. Updates 73, 101037. https://doi.org/10.1016/j.drup.2023.101037 (2024).
Wei, S. et al. Xanthohumol, a prenylated flavonoid from Hops, exerts anticancer effects against gastric cancer in vitro. Oncol. Rep. 40(6), 3213–3222. https://doi.org/10.3892/or.2018.6723 (2018).
Cao, J. et al. Influence of autologous dendritic cells on cytokine-induced killer cell proliferation, cell phenotype and antitumor activity in vitro. Oncol. Lett. 12(3), 2033–2037. https://doi.org/10.3892/ol.2016.4839 (2016).
Su, M., Hu, R., Tang, T., Tang, W. & Huang, C. Review of the correlation between Chinese medicine and intestinal microbiota on the efficacy of diabetes mellitus. Front. Endocrinol. https://doi.org/10.3389/fendo.2022.1085092 (2023).
Asgarpanah, J. & Ramezanloo, F. Chemistry, pharmacology and medicinal properties of Peganum harmala L.. Afr. J. Pharmacy Pharmacol. 6(22), 1573–1580 (2012).
Gayathiri, E. et al. Multitargeted pharmacokinetics, molecular docking and network pharmacology-based identification of effective phytocompounds from Sauropus androgynus (L.) Merr for inflammation and cancer treatment. J. Biomol. Struct. Dyn. Impact 3, 1–14. https://doi.org/10.1080/07391102.2023.2243335 (2023).
Celmar Costa Franca, T., Paula Guimaraes, A., Augusto Cortopassi, W., Alves Oliveira, A. & Castro Ramalho, T. Applications of docking and molecular dynamic studies on the search for new drugs against the biological warfare agents Bacillus anthracis and Yersinia pestis. Curr. Comput.-Aided Drug Design 9(4), 507–517 (2013).
Chen, L. et al. The roles and mechanism of m6A RNA methylation regulators in cancer immunity. Biomed. Pharmacother. 163, 114839. https://doi.org/10.1016/j.biopha.2023.114839 (2023).
Liu, H. et al. Current development of thiazole-containing compounds as potential antibacterials against methicillin-resistant staphylococcus aureus. ACS Infectious Dis. https://doi.org/10.1021/acsinfecdis.3c00647 (2024).
Zhao, Y. et al. In vitro neutralization of autocrine IL-10 affects Op18/stathmin signaling in non-small cell lung cancer cells. Oncol. Rep. 41(1), 501–511. https://doi.org/10.3892/or.2018.6795 (2019).
Zhao, J. et al. Tumor cell membrane-coated continuous electrochemical sensor for GLUT1 inhibitor screening. J. Pharm. Anal. 13(6), 673–682. https://doi.org/10.1016/j.jpha.2023.04.015 (2023).
Huang, B. et al. Babao Dan alleviates gut immune and microbiota disorders while impacting the TLR4/MyD88/NF-кB pathway to attenuate 5-Fluorouracil-induced intestinal injury. Biomed. Pharmacother. 166, 115387. https://doi.org/10.1016/j.biopha.2023.115387 (2023).
Niu, M., Guo, H., Shang, J. & Meng, X. Structural characterization and immunomodulatory activity of a mannose-rich polysaccharide isolated from bifidobacterium breve H4–2. J. Agric. Food Chem. 71(49), 19791–19803. https://doi.org/10.1021/acs.jafc.3c04916 (2023).
Zhou, Y. et al. Regulatory roles of three miRNAs on allergen mRNA expression in Tyrophagus putrescentiae. Allergy 77(2), 469–482. https://doi.org/10.1111/all.15111 (2022).
Zhang, R. et al. Recent advances of nanomaterials for intervention in Parkinson’s disease in the context of anti-inflammation. Coord. Chem. Rev. 502, 215616. https://doi.org/10.1016/j.ccr.2023.215616 (2024).
Ghasemi, K., Ghasemi, Y. & Ebrahimzadeh, M. A. Antioxidant activity, phenol and flavonoid contents of 13 citrus species peels and tissues. Pak. J. Pharm. Sci. 22(3), 277–281 (2009).
Arnao, M. B., Cano, A. & Acosta, M. The hydrophilic and lipophilic contribution to total antioxidant activity. Food Chem. 73(2), 239–244 (2001).
Bibi, Y. et al. Antibacterial activity of some selected medicinal plants of Pakistan. BMC Complementary Altern. Med. 11(1), 1–7 (2011).
George, G., Lakhani, K., & Puranam, P. What has changed? The impact of Covid pandemic on the technology and innovation management research agenda. Journal of Management Studies. (2020).
Jemal, K. Molecular docking studies of Phytochemicals of allophylus serratus Against cyclooxygenase-2 enzyme. bioRxiv, 866152, (2019)
Lipinski, C. A. Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv. Drug Delivery Rev. 101, 34–41 (2016).
Walters, W. P. Going further than Lipinski’s rule in drug design. Expert Opin. Drug Discov. 7(2), 99–107 (2012).
Kim, S. et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 49(D1), D1388–D1395 (2021).
Burley, S. K. et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 49(D1), D437 (2021).
Adejoro, I. A., Babatunde, D. D. & Tolufashe, G. F. Molecular docking and dynamic simulations of some medicinal plants compounds against SARS-CoV-2: An in silico study. J. Taibah Univ. Sci. 14(1), 1563–1570 (2020).
Acknowledgements
The authors extend their appreciation to Researchers Supporting Project number (RSP-2024 R369), King Saud University, Riyadh, Saudi Arabia.
Funding
This work was supported by National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (NRF-2021R1F1A1055482).
Author information
Authors and Affiliations
Contributions
Y.B. conceived the idea. A.K. conducted the experiment. A.A.M. and K.A.A. collected the literature review. G.M. and Y.B. provided technical expertise. I.K. and G.M. helped in statistical analysis. A.Q., S.H.Y. and Y.B. proofread and provided intellectual guidance. All authors read the first draft, helped in revision, and approved the article. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khanum, A., Bibi, Y., Khan, I. et al. Molecular docking of bioactive compounds extracted and purified from selected medicinal plant species against covid-19 proteins and in vitro evaluation. Sci Rep 14, 3736 (2024). https://doi.org/10.1038/s41598-024-54470-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-54470-6
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.