Abstract
Fritillaria cirrhosa D. Don (Liliaceae), a valuable and critically endangered medicinal herb of northwest India, including Jammu and Kashmir, grows in temperate to alpine regions of the Himalaya. It is known as the traditional herb for cardiovascular diseases, respiratory diseases, and metabolic disorders. The plant bulbs are precious and are used to cure many other health complications. The current study analysed the phytoconstituents by liquid chromatography-mass spectrometry (LC–MS) of different crude extracts (methanolic, petroleum ether, and ethyl acetate) of F. cirrhosa. The LC–MS analysis from the bulbs of F. cirrhosa yielded 88 bioactive compounds, with the vast majority having therapeutic applications. Further, determination of minimum inhibitory concentrations (MICs) by broth microdilution method of F. cirrhosa against tested bacterial and fungal pathogens showed remarkable results with MICs ranging between 6.25–200 µg/mL and 50–400 µg/mL, respectively. Subsequently, these 88 identified phytocompounds were tested for their bioactivity through ADMET prediction by SwissADME and in silico molecular docking studies. Results revealed that Peonidin might have maximum antibacterial and antifungal activity against various microbial protein drug targets among the phytochemical compounds identified. Furthermore, the highest binding affinity complex was subjected to molecular dynamic simulation (MDS) analysis using Desmond Schrodinger v3.8. The root-mean-square deviation (RMSD) graphs obtained through the molecular dynamic simulations indicated the true bonding interactions, further validated using the root-mean-square fluctuation (RMSF) graphs which provided a better understanding of the amino acids present in the proteins responsible for the molecular motions and fluctuations. To our best knowledge, this is the first description of the phytochemical constituents of the bulbs of F.cirrhosa analyzed through LC–MS, which show pharmacological significance. The in silico molecular docking and molecular dynamics study of peonidin was also performed to confirm its broad-spectrum activities based on the binding interactions with the antibacterial and antifungal target proteins. The present study results will create a way for the invention of herbal medicines for several ailments by using F. cirrhosa plants, which may lead to the development of novel drugs.
Similar content being viewed by others
Introduction
Medicinal plants have played a pivotal role in primary health care and offer a rich source of novel bioactive compounds in drug discovery and development1,2. Infectious diseases are public health problems and a significant cause of death worldwide3,4,5. Infections due to pathogenic microorganisms cause a severe concern to human health6. Increasing cases of drug resistance, unwanted side effects of existing antibiotics, and the reappearance of earlier known infections have demanded the need for new, safe and effective antimicrobial agents3,7,8,9,10. In drug development, virtual screening like drug-likeness and ADMET analysis are computational methods to find compounds that are likely to exhibit physiological activity in a short time and at a low cost using various in silico simulation methods11.
The Liliaceae family harbours a yellow Himalayan Fritillary12,13,14 that flowers yellowish to green or brownish to purple bell-shaped flowers commonly found in the alpine regions ranging from 2700–4900 m13,14, and are native to China and Indian subcontinent regions. It is widely known as Fritillaria cirrhosa D. Don. is a perennial and critically endangered medicinal herb of the Astavarga group [a group of eight medicinal herbs used in traditional medical knowledge (TMK) and the traditional medical system (TMS) in India]. Grows in open sunny slopes of temperate to alpine regions of the Himalaya15. It occurs in western temperate Himalaya from Kashmir to Kumaon from 2700 to 4035 m asl16 and from Pakistan to Uttarakhand at 2700–4000 masl17. As per the field survey conducted during the study, the species was found in the Gulmarg, Kongdori, Khillenmarg and Botapathri regions of Kashmir. According to the records of the KASH (Kashmir University Herbarium), the species has been found in the Gulmarg, Apharvat, Thajwas, Zojila, Langait, and Gurez regions of Kashmir. The bulbs of this plant are rich in bioactive compounds like sipeimine, which is used to treat many respiratory disorders. More than a hundred important bioactive phytocompounds and other significant chemical compounds have identified, isolated and extracted till date from F. cirrhosa18. These components include alkaloids like peiminine, peimine, peimisine, imperialine etc.19,20,21. Chinese medicinal practitioners have employed this plant to implicate its medicinal and therapeutic potency22. It's a bitter tonic and gastrointestinal stimulant that relieves fevers and urinary tract infections and is a medicine for 80 diseases23. It's used as a refrigerant, diuretic, galactagogue, expectorant, and aphrodisiac, among other things. It's used to treat rheumatism, tuberculosis, and haematemesis. It also relieves discomfort in pregnant women, encourages the growth of flesh, and relieves various aches and symptoms24. The alkaloids found in the bulbs of F. cirrhosa have been shown to have hypotensive21,25, anti-inflammatory21,26, antitumor21,25,27, antitussive, expectorant21,26,28, and antiasthmatic activities21,28.
These findings support the hypothesis that F. cirrhosa is a valuable medicinal plant. Although the extracts of F. cirrhosa were traditionally utilized and therefore extensively studied and reported for various pharmacological activities. Since the chemistry and antimicrobial studies of the F. cirrhosa growing in Jammu & Kashmir seems not to have been worked out, we carried out the present studies to characterize the various extracts and investigate its antimicrobial activity through molecular docking studies, ADMET analysis and in vitro approaches for the first time. Herein, we report the antimicrobial activity of three different extracts against various bacterial and fungal strains.
Methodology
Collection of plant material
F. cirrhosa bulbs were collected from four locations (viz Razdhan top, Sadhna Pass, Tangdhar Main, Apharvat Gulmarg) in 2020 of Jammu and Kashmir. Geographical coordinates of the site sample are mentioned in (Table 1). The plant was identified by Akhtar Malik, Curator and Taxonomist at the University of Kashmir's Department of Botany.
Preparation of extracts
Petroleum ether, ethyl acetate, and methanol were chosen as extraction solvents based on their polarity index. Finely ground powdered plant material (5 g) was extracted in the solvents as mentioned above. To maintain the natural antimicrobial activity in a sample extract and allow maximum extraction, the mixture was stored on an incubator rotatory shaker at (200 rpm) 25 °C for 48 h. After filtering the extracts through Whatman filter paper (No. 1), the filtrate was centrifuged for 15 min at 8000 rpm at 12 °C to obtain a clear supernatant. The extracts produced in various solvents were evaporated under reduced pressure in a rotatory evaporator and diluted to obtain a stock solution with a concentration of 10 mg/mL, which was stored in falcon tubes coated in aluminium foil at 4 °C in the refrigerator for later use29.
Liquid chromatography–mass spectrometry (LC/MS)
Nexera UHPLC with a quaternary pump, central degassing unit, Autosampler, and DAD unit was used for the LC–MS analysis. Elution was carried out at a rate of 1 mL/min with petroleum ether, ethyl acetate and methanol as solvents. After ultrasonication, all solvents were passed through 0.45 µm nylon paper. At 270 nm, the chromatograms were examined, and the findings were obtained using lab-developed software30. The plant extracts were formulated using petroleum ether, ethyl acetate, methanolic, and bioanalysis to identify important metabolites and phytocompounds. The presence of 88 phytocompounds were ascertained by executing LC–MS analysis on these extracts.
Bacterial strain and culture preparation
The microbes were procured from IMTECH's Gene Bank in Chandigarh and American Gene Bank. For antibacterial and antifungal activity, six bacterium strains and three fungus strains were investigated (Escherichia coli MTCC443, Klebsiella pneumonia MTCC19, Microbacterium luteus, Streptococcus pneumonia MTCC 655, Haemophilus influenzae MTCC 3826, Neisseria mucosa MTCC 1722 and Candida albicans ATCC 24433, Candida glabrata ATCC2001, Candida Parapsilosis ATCC90018).
All the cultures were maintained at 37 °C for better growth in the incubator. For each experiment, the prepared aliquots were thawed and sub-cultured in broth with 10% ADC (albumin-dextrose-catalase Himedia, India), 0.2% Glycerol (Merck, India), and 0.05% Tween 80 (Merck, India) and cultures were grown till stationary phase and further processed for other experiments.
Minimum inhibitory concentration by broth dilution method
Minimum inhibitory concentration in the present study was performed through Muller Hinton broth dilution method. In the present study, 10% ADC was supplemented in broth followed by performing a two-fold serial concentration from 0.00 to 0.20 µg/mL of antibiotics, including ciprofloxacin and amphoterecin-B in different combinations with fungal and bacterial cultures. Moreover, each inoculated plate was kept in an incubator at 37 °C for 24 to 48 h. To monitor the growth of cells and observe growth inhibition, the minimum inhibitory concentration of the drug was noted. MIC was considered as the lowest concentration of an antimicrobial at which no balanced growth was observed. In addition, colony-forming units were, measured on day 28th of incubation at 37 °C for better use.
In terms of drug interaction, the microbial cultures were grown in various concentrations of ciprofloxacin and amphotericin B in multiple combinations. The antimicrobials were added to fabricate 8 × MIC for both the antimicrobials followed by two fold dilutions leading to 1/64 × MIC concentration ranges of each drug both alone and in combination were assured31.
Ligand and receptor preparations
The molecular docking analysis was carried out for all the selected phytocompounds from the LC–MS data fractions. Chemical structures of phytocompounds retrieved from PubChem database in SDF format and converted into protein data bank (PDB) format using PyMol Version 3.0. Software. The selected targets for this study were based on previous studies, and all the 3D crystal structures were fetched from (PDB) in PDB format. The structural information of the Peonidin identified from the Fritillaria cirrhosa was also retrieved from the PubChem database in SDF format and converted into PDB format using an open babel server. Peonidin is one of the compounds identified from LC–MS analysis. It exhibits apoptotic effects in the breast cancer cells32.
Peonidin is an anthocyanide that can be easily eliminated from the human body. Anthocyanins (ACNs), including peonidin, are natural bioactive compounds with many pharmacological effects: antioxidant, anti-inflammatory, prevention of age-related chronic diseases: cardiovascular disease (CVD), cancers, neurodegenerative, and eye-related diseases. ACNs also have antiviral properties. Recent in vitro studies have shown that they can inhibit the replication of viruses such as herpes simplex, parainfluenza virus, syncytial virus, HIV, rotavirus, and adenovirus33. The broad spectrum of pharmacological properties supported by preclinical and clinical evidence associated with low toxicity make their pharmacotherapeutic use very attractive. Peonidin was selected as an appropriate ligand in the current study based on its pharmacological properties. Compounds with antimicrobial activities disable bacteria by targeting key bacterial metabolism components, including dihydropterote synthase (DHPS), penicillin binding protein (PBP), elongation factor Tu, 1,3 beta glycan, ABC transporter and beta-tubulin . These six key proteins play a critical role in the life cycle of fungi and bacteria. Penicillin-binding proteins (PBPs) are membrane-associated proteins that play a vital role in forming cell wall34,35. As previously indicated, antibiotics target the dihydropteroate synthase enzyme36. This enzyme participates in the folate synthesis pathway, directly connected to nucleic acid synthesis37. Inhibiting this enzyme with a strong antibiotic will have a more profound and irreversible effect on the bacterium's protein production. EF-Tu's primary role is to transport amino-acylated tRNAs to the ribosome. Since the 1970s, antibiotics have targeted EF-Tu as a therapeutic38. ABC transporters (ATP-binding cassette) are found in all three domains of life and mediate transmembrane transport of a wide range of substrates, including medicines, proteins, carbohydrates, amino acids, ions and carbohydrates. Glucans have long been known to have immunomodulatory effect39. Tubulin are the protein subunits of microtubules which helps in cell division of prokaryotic and eukaryotic organisms40. In view of this, the molecular docking study was carried out to examine the binding interactions of peonidin with these microbial drug targets. All the targeted receptors were fetched from the protein data bank (PDB) in the PDB format and subjected for purification and refinement. All the receptors were prepared for molecular docking analysis using Biovia Discovery Studio and eliminate undesirable ligands, molecules of water, and other impurities from all of the proteins involved in the study.
Purification and refinement of proteins and ligands
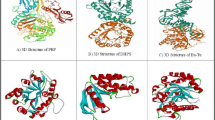
Biovia Discovery studio was used to eliminate undesirable ligands, molecules of water, and other impurities from all of the proteins involved in the study. Polar hydrogens were added to the protein during the preparation process for improved interactions, followed by Kollman charges. The 2D structure and 3D structure of peonidin were retrieved using the PubChem database (PubChem CID: 5281708). The 3D structures of dihydropteroate synthase (DHPS), penicillin-binding protein (PBP), elongation factor Tu, 1,3 beta glycan, ABC transporter and beta-tubulin were retrieved from the Protein Data Bank (Fig. 1).
Molecular docking studies
The docking calculation and algorithms to reproduce the pocket binding residues was verified employing co-crystal ligands. In the current study, we have used the Lamarckian Genetic Algorithm in Autodock Vina 4.2 to initiate the docking analysis with the standard parameters. Furthermore, all the transformation were clustered by considering the 4.0A RMSD and the most favourable binding poses was selected depending on the lowest free energy and inhibition constant. The molecular docking studies were performed on all selected phytocompounds from the LC–MS data sections. Chemical structures of phytocompounds were obtained in SDF format from the PubChem database and were translated to PDB format with PyMol Version 3.0. The targets for this investigation were chosen based on the evidence from past research, and all 3D crystal structures were collected in .pdb format from the Protein Data Bank. Based on the isolation modes and species-specific variants, the crystal structures of proteins such as dihydropterote synthase, penicillin-binding protein, and elongation factor Tu (Ef-Tu), 1,3 beta glycan, ABC transporter and beta-tubulin were retrieved.
In-silico drug-likeness and toxicity predictions
Drug-likeness of compounds was predicted based on an already established concept by Lipinski et al.41. The structures of all some pharmacologically important compounds were converted to their canonical simplified molecular-input line-entry system (SMILE). They submitted to the SwissADME and PreADMET tool to estimate in silico pharmacokinetics, such as the number of hydrogen donors, hydrogen acceptors, rotatable bonds, and total polar surface area of a compound. These compound’s organ toxicities and toxicological endpoints were predicted using PreADMET and OSIRIS Property42. The selection of compounds as drug candidates was determined by a drug score parameter. The higher the drug score value, the higher the compound's chance of being considered a drug candidate43.
Molecular dynamic simulation
The Desmond Schrodinger package was employed for the molecular dynamic simulation with the highest binding affinity docked complex to analyse the docked calculation and interpretation. The analysis indulged simulation run for 100 ns to check the stability of structures. Desmond's inbuilt parameters were also used for protein preparation and bond orders. In the case of bond orders, the addition of hydrogen binds, and filling of missing residues are also optimized during the initial process. OPLS force field was used to initiate the builder system for the standard simulation grid box. And the conformation were retrieved in the form of simulation trajectories which could be analyzed using Root mean square deviation and root mean square fluctuation graphs through hydrogen bond analysis and radius of gyration plots.
Results
Preliminary phytochemical screening
The phytochemical study of various extracts from the bulbs of F. cirrhosa revealed a variety of phytochemicals. The essential constituents present in petroleum ether, ethyl acetate and methanolic extracts as represented in Table 2.
Chemical composition
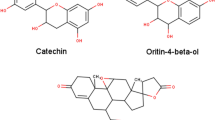
The separation of individual components presents in a solvent-based on their mass/charge ratio is the working principle behind Liquid Chromatography-Mass Spectroscopy (LC–MS). The chemical composition of the extracts from the F. cirrhosa, analyzed by LC–MS, are represented in (Table 3; Fig. 2). The LC–MS total ion chromatograms of different extracts of F. cirrhosa are shown in (Figs. 3, 4, 5). The solvent extraction carried out by Soxhlet extraction was subjugated to LC–MS analysis to obtain 88 important bioactive phytocompounds (Supplementary data file). A total of 22 phytocompounds was obtained using the methanolic extract etc. d-Glucose-6-phosphate disodium salt, 3-Hydroxy-dl-kynurenine, Leucylleucyltyrosine, and Ononin are a few compounds procured through the methanolic extract. Linarin, 1,6-Anhydro-beta-d-glucose, O-Phosphoryl ethanolamine were significant proportions among the 28 phytocompounds obtained through the petroleum ether extract. Peonidin, 2,5,7,8-Tetramethyl-2-(4,8,12-trimethyltridecyl)-6-chromanol Acetate, Arachidonic acid, and 19 other compounds were derived using the ethyl acetate extract.
Minimum inhibitory concentration
In terms of novel antimicrobial drugs, we have investigated the antimicrobial compounds extract from F. cirrhosa that showed significant inhibitory activity shown in Table 4 and observed to be non-toxic. The present study elaborates the detailed antimicrobial activity and in silico study for better validation.
In the present study, MICs of standard antimicrobial drug targets ciprofloxacin and amphotericin-B through broth dilution were 0.625, 1.25, 0.039, 0.625, 1.25, 3.12, 1.25, 2.5, and 2.5 µg/mL, respectively. The minimum inhibitory concentration of different plant extracts showed better outcomes than co-crystals shown in Table 4.
Molecular docking analysis
The LC–MS analysis revealed that F. cirrhosa bulb extracts contained 88 bioactive compounds (Table 3). These phytocompounds were analyzed for activities against bacterial and fungal target proteins. The docking studies were carried out for phytoligands using the AutoDock Vina programme to elucidate the binding affinities to the target proteins. In general, among the 88 phytocompounds. All the six microbial drug targets related protein structures were docked against best-screened ligand peonidin obtained after ADME screening based on different parameters like solubility, toxicity, absorption, molecular weight and excretion. Docking was performed using AutoDock Vina with Chimera Plugin. Subsequently, the best-docked complex was screened based on their binding energy. Bonding between ligand and target protein were visualized using Biovia discovery studio. 2D-interaction diagrams showing different bonds formed between ligand and target protein were also visualized using Biovia discovery studio. Peonidin (2-(4-hydroxy-3-methoxyphenyl) chromenylium-3,5,7-triol) exhibited the best binding conformations with the lowest binding energy values with bacterial (− 8.2 kcal/mol), fungal (− 8.2 kcal/mol) target proteins (Tables 5 and 6). Our findings are supported by a previous study that showed that the lower the binding energy score, the better the protein–ligand binding stability was identified44. The peonidin formed the most excellent ligand–protein complexes with six tested microbial target proteins compared with other compounds. According to the binding energies, the docking results of peonidin with target proteins were ranked. The docking results of peonidin with the proteins, such as dihydropterote synthase, penicillin-binding protein, elongation factor Tu, 1, 3 beta glycan, ABC transporter and beta-tubulin confirmed that the ligand has a higher affinity for penicillin-binding protein, which is a key regulator in cell wall synthesis and maintenance (Fig. 6). In comparison to ciprofloxacin, the extracted peonidin natural molecule comparatively showed same docking efficiency with PBP within the binding pocket. The docked structure was imaged to illustrate the ligand interactions with significant amino acids TYR A:561, ILE A:371, ASN A:397, PHE A:450, and GLN A:452 through Vander Waal forces as well as hydrogen bonding (Fig. 6). The binding affinity of the selected compound against antibacterial and antifungal proteins are represented in (Tables 5, 6, 7, 8).
(a) The 2D and 3D intermolecular contact between ciprofloxacin and Pencillin binding protein. Chemical structures were drawn by ChemDraw Pro 16.0 Suite (PerkinElmer, USA) and analyzed by the Discovery studio visualizer (BIOVIA Discovery studio 2020 Client). (b) The 3D and 2D intermolecular contact between A) Dihydropteroate synthase B) Elongation factor Tu and C) Penicillin Binding Protein D) 1,3-Betaglycan E) Beta-tubulin F) ABC transporter with peonidin . Chemical structures were drawn by ChemDraw Pro 16.0 Suite (PerkinElmer, USA) and analyzed by the Discovery studio visualizer (BIOVIA Discovery studio 2020 Client).
Molecular dynamic simulation
The best-hit compound was considered based on the binding affinity and transformation for molecular dynamic studies to analyze the stability of the compound in contrast to the protein of our interest. For simulation studies, the Desmond Schrodinger suite program was used for receptor complexes followed by the RMSD of protein exhibited a stable trajectory during the dynamic run for 100 nano seconds studies. Simulation of 100 nano seconds indicates significant conversions and stable conformations when comparing protein–ligand RMSD, as shown in Fig. 7. Regarding dynamic studies, RMSD exposed a fluctuated trajectory till 30 nano seconds, and there are fewer fluctuations between ligand and protein conformations (Fig. 7). Overall, both the receptor and ligand RMSD was observed in a stable format and can be seen in (Figs. 7, 8, 9, 10). In addition, all the amino acid interactions of our complexes were also analyzed accordingly, as well as the unique hydrogen bonds noted like TYR 561, ILE 371, ASN 397, and so on.
Protein–ligand contacts (VAL 72, ARG 254, THR 257, LYS 259, THR 275, ALA 279, GLY 28, LYS 283, LYS 285, SER 337, LYS 340, ARG 372, TRP 374, ASN 377, GLY 378, LYS 380, ARG 380, HIS 354, SER 394, ASNB 395, PHE 397, GLY 400 , ASN 452, ASN 487, THR 488, ARG 490, ASP 492, PRO 521, VAL 522, TYR 523, GLY 524, TYR 525, MET 526, TYR 527, ASN 525, HIS 529, SER 531, TYR 531, LYS 532, SER 534, GLY 548, THR 549, GLY 550, VAL 552, LYS 583, THR 565, TYR 566, PHE 568, GLU 570, HIS 593, TYR 594, SER 595, GLY 596, ILE 597, GLN 598, GLU 599, ASN 602, GLN 608, GLN 632, PRO 633, PRO 635, ASN 677).
Discussion
F. cirrhosa is a versatile medicinal plant with numerous biological properties in its nature. In the present study, the investigation of petroleum ether, ethyl acetate, and methanol extracts from bulbs of F. cirrhosa revealed the presence of various phytoconstituents, including flavonoids and carbohydrates saponins, phenols, alkaloids, steroids, and terpenoids. These bioactive phytoconstituents could be responsible for the therapeutic ability of various extracts of F. cirrhosa. The analysis was carried out by liquid chromatography-mass spectrometry (LC–MS), one of the most widely used techniques for separating phyto-constituents. The LC–MS investigation of F. cirrhosa bulb extracts revealed the presence of 88 phytochemical compounds, which could contribute to the medicinal properties of this plant species14. Solasodine (1S,2S,4S,5'R,6R,7S,8R,9S,12S,13R,16S)5',7,9,13tetramethylspiro[5oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-18-ene-6,2'-piperidine]-16-ol) were extracted from Solanum dulcamara and evaluated for antimicrobial activity against the test bacteria like Staphylococcus aureus, Enterobacter aerogenes, Escherichia coli45,46. Linarin (5-hydroxy-2-(4-methoxyphenyl)-7-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxymethyl]oxan-2-yl]oxychromen-4-one) a bioactive flavone glycoside has been characterized abundantly in the Cirsium, Micromeria, and Buddleja species and exhibited promising activities particularly, the remedial effects on central nervous system (CNS) disorders47. Similarly, puerarin (7-hydroxy-3-(4-hydroxyphenyl)-8-[(2S,3R,4R,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]chromen-4-one) is the major bioactive ingredient isolated from the root of the Pueraria lobata has been widely used in the treatment of cardiovascular and cerebrovascular diseases, diabetes and diabetic complications, osteonecrosis, Alzheimer’s disease, Parkinson’s disease, endometriosis, and cancer48. Likewise, Orientin isolated from different medicinal plants and possess promising pharmacological activities, which include antioxidant, antiaging, antiviral, antibacterial, anti-inflammation, vasodilatation and cardioprotective, radiation protective, neuroprotective, antidepressant-like, antiadipogenesis, and antinociceptive effects49. There are many reports on peonidin which exhibits apoptotic effects in the breast cancer cells and therapeutic applications25,50. Furthermore, alkaloids like sinpeinine A (17), imperialine-3-β-glucoside (18), imperialine (7) and 3β-acetylimperialine have been reported from Bulbus F. cirrhosa14. From the above evidence, it can be elucidated that F. cirrhosa consists of the enormous potential of pharmacological constituents therapeutic phytocompounds responsible for various pharmacological actions. This is the first report on LC–MS investigation of F. cirrhosa bulb extracts to the best of our knowledge. Furthermore, the present study investigated the in vitro antimicrobial activity against various bacterial and fungal strains. The study focused on the gram-positive bacteria and gram-negative bacteria, which shows the significant efficiency. Among various fungal and bacterial strains such as E. coli, K. pneumonia, M. luteus, S. pneumonia, H. influenzae, N. mucosa and C. albicans, C. glabrata, C. Parapsilosis were observed to be more susceptible to the plant extracts with the better minimum inhibitory concentration values (Table 4) compare to co- crystals MICs (0.625, 1.25, 0.039, 0.625, 1.25, 3.12, 1.25, 2 0.5, and 2.5) at a combinational concentration. In addition, it is also mentioned to that, the therapeutic herbs have different mechanisms that reflects identical antimicrobial mode of therapeutics in contrast to antimicrobial agents10,51. According to molecular docking, peonidin with 6 different proteins (dihydropterote synthase, penicillin binding protein, elongation factor Tu, 1,3 beta glycan, ABC transporter and beta-tubulin) exposed that the peonidin has a better interaction and binding score with penicillin binding protein as compared to other phytocompounds. The docking study results also showed that various energy sources are consistent and contribute to the overall strength of the binding interactions of peonidin for each target protein. After docking, computational molecular dynamic simulation studies revealed that various amino acids were observed to have interacted with peonidin, which is requisite for the synthesis of purines and pyrimidine nucleotides. Most of the interacted amino acids were bound with the hydrogen bonding and Vander Wall forces, whereas, the docking outcome with dihydropteroate synthase exposed the interrelated binding affinity with targeted receptors. The trajectory of molecular dynamic simulation was completed for 100 nano seconds and the RMSD and RMSF graph values were analyzed. The constant increase in the RMSD values with respect to time indicates that the protein serves from its native conformation regularly. The higher RMSF was analyzed in the study where strong hydrogen bonding was noted with mostly polar amino acids having maximum peak.
Conclusion
The present investigation focused on identifying various bioactive compounds from the bulb extracts of F. cirrhosa for the first time by LC–MS analysis. These compounds are responsible for the different therapeutic and pharmacological properties. We have also provided evidence of bulb extracts of F. cirrhosa for its antimicrobial activity. Furthermore, the present study also explained the role of the peonidin compound against antimicrobial proteins such as dihydropteroate synthase (DHPS), penicillin-binding protein (PBP), elongation factor-Tu (Eu-Tu), 1,3 β-glycan, ABC transporter, and beta-tubulin. Among all the bacterial and fungal species, peonidin showed the best docking score against PBP and was then subjected to dynamic simulation studies to investigate the stability and RMSD and RMSF values concerning protein–ligand transformations. Using the peonidin compound may enable us to develop an effective drug against pathogenic bacteria and fungal diseases. In addition, ADMET (drug-likeness) studies showed the highest drug-likeness properties of the studied compound, which suggests that the peonidin compound can act as a promising microbail drug candidate. Further investigations to determine its bioactivity, toxicity profile, and clinical studies are necessary for broad-spectrum drug discovery.
Abbreviations
- ATCC:
-
American Type Culture Collection Centre
- CFU:
-
Colony-forming units
- CIP:
-
Ciprofloxin
- MIC:
-
Minimum inhibitory concentration
- MTCC:
-
Microbial Type Culture Collection Centre
- LC–MS:
-
Liquid chromatography coupled with mass spectroscopy
- UHPLC:
-
Ultra-high-performance liquid chromatography
- CBT:
-
Centre for biodiversity and taxonomy
- IMTECH:
-
Institute of microbial technology
- PBP:
-
Penicillin binding protein
- PAMPs:
-
Pathogen associated molecular pattern
- PDB:
-
Protein Data Bank
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- MD:
-
Molecular docking
- MDS:
-
Molecular dynamic simulation
References
Mir, M. A., Hamdani, S. S., Sheikh, B. A. & Mehraj, U. Recent advances in metabolites from medicinal plants in cancer prevention and treatment. Curr. Immunol. Rev. 15, 185–201 (2019).
Bhat, B. A., Nisar, S., Sheikh, B. A., Mir, W. R. & Mir, M. A. Antioxidants (natural and synthetic) screening assays: An overview. Bentham Br. Biomed. Pharmacother. Oxid. Stress Nat. Antioxid., 105 (2021).
Sheikh, B. A. et al. Development of new therapeutics to meet the current challenge of drug-resistant tuberculosis. Curr. Pharm. Biotechnol. 22, 480–500 (2021).
Sheikh, B. A. et al. Nano-drug delivery systems: Possible end to the rising threats of tuberculosis. J. Biomed. Nanotechnol. 17, 2298–2318 (2021).
Mir, M. A. et al. in Applications of Nanomaterials in Agriculture, Food Science, and Medicine 229–251 (IGI Global, 2021).
Qadri, H., Shah, A. H. & Mir, M. Novel strategies to combat the emerging drug resistance in human pathogenic microbes. Curr. Drug Targets 22, 1424–1436 (2021).
Miethke, M. et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 5, 726–749 (2021).
Daszak, P. Global trends in emerging infectious diseases. Nature 451, 990–993 (2008).
Cassell, G. H. & Mekalanos, J. Development of antimicrobial agents in the era of new and reemerging infectious diseases and increasing antibiotic resistance. JAMA 285, 601–605 (2001).
Bhat, B. A. et al. In vitro and in silico evaluation of antimicrobial properties of Delphinium cashmerianum L., a medicinal herb growing in Kashmir. India. J. Ethnopharmacol. 291, 115046 (2022).
Ekins, S., Mestres, J. & Testa, B. In silico pharmacology for drug discovery: Methods for virtual ligand screening and profiling. Br. J. Pharmacol. 152, 9–20 (2007).
Beetle, D. E. A monograph of the North American species of Fritillaria. Madroño 7, 133–159 (1944).
Roguz, K. et al. Functional diversity of nectary structure and nectar composition in the genus Fritillaria (Liliaceae). Front. Plant Sci. 9, 1246 (2018).
Rashid, I. & Yaqoob, U. Traditional uses, phytochemistry and pharmacology of genus Fritillaria—A review. Bull. Natl. Res. Cent. 45, 1–37 (2021).
Chauhan, R. S. et al. Morpho-biochemical variability and selection strategies for the germplasm of Fritillaria roylei Hook (Liliaceae)—An endangered medicinal herb of western Himalaya, India. J. Plant Breed. Crop Sci. 3, 430–434 (2011).
Khare, C. P. Indian Medicinal Plants: An Illustrated Dictionary 699–700 (Springer, 2007).
Prakash, K. & Nirmalaa, A. A review on conservation of endangered medicinal plants in India. Int. J. Ethnomed. Pharmacol. Res. 1, 21–33 (2013).
Kumar, P., Partap, M. & Warghat, A. R. in Himalayan Medicinal Plants 57–66 (Elsevier, 2021).
Mondal, A., Gandhi, A., Fimognari, C., Atanasov, A. G. & Bishayee, A. Alkaloids for cancer prevention and therapy: Current progress and future perspectives. Eur. J. Pharmacol. 858, 172472 (2019).
Zhang, Q.-J., Zheng, Z.-F. & Yu, D.-Q. Steroidal alkaloids from the bulbs of Fritillaria unibracteata. J. Asian Nat. Prod. Res. 13, 1098–1103 (2011).
Wang, D. et al. Optimization of extraction and enrichment of steroidal alkaloids from bulbs of cultivated Fritillaria cirrhosa. BioMed Res. Int. 2014 (2014).
Kumar, P., Acharya, V. & Warghat, A. R. Comparative transcriptome analysis infers bulb derived in vitro cultures as a promising source for sipeimine biosynthesis in Fritillaria cirrhosa D. Don (Liliaceae, syn. Fritillaria roylei Hook.)-High value Himalayan medicinal herb. Phytochemistry 183, 112631 (2021).
Sultan, R., Wani, M. A. & Nawchoo, I. A. Unabated loss of medicinal plant diversity in Himalaya: A serious socio-economic concern and urgency to salvage whatever is left. Glob. Adv. Res. J. Med. Plants 2, 012–021 (2013).
Dhyani, A., Nautiyal, B. P. & Nautiyal, M. C. Importance of Astavarga plants in traditional systems of medicine in Garhwal, Indian Himalaya. Int. J. Biodivers. Sci. Ecosyst. Serv. Manag. 6, 13–19 (2010).
Wang, D.-D. et al. In vitro and in vivo antitumor activity of Bulbus Fritillariae Cirrhosae and preliminary investigation of its mechanism. Nutr. Cancer 66, 441–452 (2014).
Wang, D. et al. Antitussive, expectorant and anti-inflammatory alkaloids from Bulbus Fritillariae Cirrhosae. Fitoterapia 82, 1290–1294 (2011).
Wang, D. et al. Antitumor effects of Bulbus Fritillariae cirrhosae on Lewis lung carcinoma cells in vitro and in vivo. Ind. Crops Prod. 54, 92–101 (2014).
Wang, D. et al. Antitussive, expectorant and anti-inflammatory activities of four alkaloids isolated from Bulbus of Fritillaria wabuensis. J. Ethnopharmacol. 139, 189–193 (2012).
Abubakar, A. R. & Haque, M. Preparation of medicinal plants: Basic extraction and fractionation procedures for experimental purposes. J. Pharm. Bioallied Sci. 12, 1 (2020).
Seo, C.-S. & Shin, H.-K. Quality assessment of traditional herbal formula, Hyeonggaeyeongyo-tang through simultaneous determination of twenty marker components by HPLC–PDA and LC–MS/MS. Saudi Pharm. J. 28, 427–439 (2020).
Rather, M. A. et al. In vitro antimycobacterial activity of 2-(((2-hydroxyphenyl) amino) methylene)-5, 5-dimethylcyclohexane-1, 3-dione: a new chemical entity against Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 52, 265–268 (2018).
Kwon, J. Y., Lee, K. W., Hur, H. J. & Lee, H. J. Peonidin inhibits phorbol-ester–induced COX-2 expression and transformation in JB6 P+ cells by blocking phosphorylation of ERK-1 and-2. Ann. N. Y. Acad. Sci. 1095, 513–520 (2007).
Mohammadi Pour, P., Fakhri, S., Asgary, S., Farzaei, M. H. & Echeverría, J. The signaling pathways, and therapeutic targets of antiviral agents: Focusing on the antiviral approaches and clinical perspectives of anthocyanins in the management of viral diseases. Front. Pharmacol. 10, 1207 (2019).
Miyachiro, M. M., Contreras-Martel, C. & Dessen, A. Penicillin-binding proteins (PBPs) and bacterial cell wall elongation complexes. Macromol. Protein Complex. II Struct. Funct., 273–289 (2019).
Macheboeuf, P. et al. Active site restructuring regulates ligand recognition in class A penicillin-binding proteins. Proc. Natl. Acad. Sci. 102, 577–582 (2005).
Potshangbam, A. M., Rathore, R. S. & Nongdam, P. Discovery of sulfone-resistant dihydropteroate synthase (DHPS) as a target enzyme for kaempferol, a natural flavanoid. Heliyon 6, e03378 (2020).
Hampele, I. C. et al. Vol. 268 21–30 (Elsevier, 1997).
Harvey, K. L., Jarocki, V. M., Charles, I. G. & Djordjevic, S. P. The diverse functional roles of elongation factor Tu (EF-Tu) in microbial pathogenesis. Front. Microbiol. 10, 2351 (2019).
Novak, M. & Vetvicka, V. β-glucans, history, and the present: Immunomodulatory aspects and mechanisms of action. J. Immunotoxicol. 5, 47–57 (2008).
Michalkova, R., Mirossay, L., Gazdova, M., Kello, M. & Mojzis, J. Molecular mechanisms of antiproliferative effects of natural chalcones. Cancers 13, 2730 (2021).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. In vitro models for selection of development candidatesexperimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25 (1997).
Oduselu, G. O., Ajani, O. O., Ajamma, Y. U., Brors, B. & Adebiyi, E. Homology modelling and molecular docking studies of selected substituted Benzo [d] imidazol-1-yl) methyl) benzimidamide scaffolds on Plasmodium falciparum adenylosuccinate lyase receptor. Bioinform. Biol. Insights 13, 1177932219865533 (2019).
Behrouz, S. et al. Design, synthesis, and in silico studies of novel eugenyloxy propanol azole derivatives having potent antinociceptive activity and evaluation of their β-adrenoceptor blocking property. Mol. Divers. 23, 147–164 (2019).
Teli, M. K., Kumar, S., Yadav, D. K. & Kim, M. H. In silico identification of prolyl hydroxylase inhibitor by per-residue energy decomposition-based pharmacophore approach. J. Cell. Biochem. 122, 1098–1112 (2021).
Kumar, R., Khan, M. I. & Prasad, M. Solasodine: A perspective on their roles in health and disease. Res. J. Pharm. Technol. 12, 2571–2576 (2019).
Jayakumar, K. & Murugan, K. Solanum alkaloids and their pharmaceutical roles: A review. J. Anal. Pharm. Res. 3, 00075 (2016).
Mottaghipisheh, J. et al. Linarin, a glycosylated flavonoid, with potential therapeutic attributes: A comprehensive review. Pharmaceuticals 14, 1104 (2021).
Zhou, Y. X., Zhang, H. & Peng, C. Puerarin: A review of pharmacological effects. Phytother. Res. 28, 961–975 (2014).
Lam, K. Y., Ling, A. P. K., Koh, R. Y., Wong, Y. P. & Say, Y. H. A review on medicinal properties of orientin. Adv. Pharmacol. Sci. 2016 (2016).
Salehi, B. et al. The therapeutic potential of anthocyanins: Current approaches based on their molecular mechanism of action. Front. Pharmacol. 11, 1300 (2020).
Sheikh, B. A., Bhat, B. A., Ahmad, Z. & Mir, M. A. Strategies employed to evade the host immune response and the mechanism of drug resistance in Mycobacterium tuberculosis: In search of finding new targets. Curr. Pharm. Biotechnol. (2021).
Acknowledgements
Authors would like to acknowledge Molecular Biology Laboratory, Division of Veterinary Biochemistry, SKUAST-K, Srinagar, for their assistance in carrying out antimicrobial activity. Mustafa F Alkhanani would like to express his gratitude to AlMaarefa University, Riyadh, KSA for providing funding (TUMA-2021-53).
Funding
Funding was provided by Science and Engineering Research Board (SERB), Department of Science and Technology, Govt of India vide Grant No. TAR/001213/2018 to Dr Manzoor Ahmad Mir.
Author information
Authors and Affiliations
Contributions
M.A.M., conceptualization, designed research work, interpreted the data, B.A.B., W.R.M. conducted the experiments, analysed the data, wrote the manuscript; M.A.M., M.A., B.A.B., B.A.S. and W.R.M. analysed the data, reviewed, edited the manuscript and approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bhat, B.A., Mir, W.R., Sheikh, B.A. et al. Metabolite fingerprinting of phytoconstituents from Fritillaria cirrhosa D. Don and molecular docking analysis of bioactive peonidin with microbial drug target proteins. Sci Rep 12, 7296 (2022). https://doi.org/10.1038/s41598-022-10796-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10796-7
This article is cited by
-
Chromatographic Fingerprinting of Cacao Pod Husk Extracts (Theobroma cacao L.): Exploring Antibacterial, Antioxidant, and Antidiabetic Properties with In Silico Molecular Docking Analysis
Applied Biochemistry and Biotechnology (2024)
-
Antimicrobial resistance: new insights and therapeutic implications
Applied Microbiology and Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.