Abstract
Reproductive success requires the development of viable oocytes and the accurate segregation of chromosomes during meiosis. Failure to segregate chromosomes properly can lead to infertility, miscarriages, or developmental disorders. A variety of factors contribute to accurate chromosome segregation and oocyte development, such as spindle assembly and sister chromatid cohesion. However, many proteins required for meiosis remain unknown. In this study, we aimed to develop a screening pipeline for identifying novel meiotic and fertility genes using the genome of Drosophila melanogaster. To accomplish this goal, genes upregulated within meiotically active tissues were identified. More than 240 genes with no known function were silenced using RNA interference (RNAi) and the effects on meiosis and fertility were assessed. We identified 94 genes that when silenced caused infertility and/or high levels of chromosomal nondisjunction. The vast majority of these genes have human and mouse homologs that are also poorly studied. Through this screening process, we identified novel genes that are crucial for meiosis and oocyte development but have not been extensively studied in human or model organisms. Understanding the function of these genes will be an important step towards the understanding of their biological significance during reproduction.
Similar content being viewed by others
Introduction
Fertility requires both a successful meiosis to provide balanced genetic complement to offspring, and several developmental processes to make viable zygotes. During meiosis, germ line cells undergo a single round of genome duplication followed by two consecutive chromosomal divisions prior to fertilization. The meiotic process is highly regulated by multiple cellular structures and protein complexes, but the processes are error-prone, especially in oogenesis. In humans, chromosome segregation errors during oocyte meiosis increase with age. This increase could be related to the unique features of oocytes, such as the extended meiotic arrest, the absence of centrosomes, and possibly other endogenous and exogenous factors1. Failure to produce high-quality gametes leads to infertility, spontaneous abortions, and birth defects2. Many genes have been identified that are responsible for accurate meiotic divisions and regulate discrete cell-cycle phases, meiotic events, and gamete viability3,4,5,6. Nevertheless, we still lack a complete picture of the proteins that control key processes in meiosis, such as chromosome cohesion, chromosome biorientation, and spindle assembly.
In addition to meiosis, defects in numerous developmental processes can also lead to infertility, such as genes required for germline development7. Furthermore, in most animals, maternally derived gene products regulate the early events of embryogenesis8. These include genes required for egg activation, early mitotic divisions, and the maternal-to-zygotic transition9,10,11. Elimination of these genes in the mother can lead to loss of fertility. It is likely that many additional factors are responsible for the loss of female fertility in humans, although few have been identified5,12.
Drosophila is a simple but vital genetic model for identifying and understanding the function of genes required for germline and embryonic development and meiosis13,14. Despite the differences between Drosophila and human physiology, the homologs of many human genetic disease loci show selective expression in Drosophila tissues analogous to the affected human tissues15. Additionally, the ability to produce a large number of offspring in a short period of time makes Drosophila a powerful system for gene discovery and studying potential disease-causing genes16,17.
In Drosophila, many well-studied genes required for meiosis are upregulated within the ovary, such as components of the synaptonemal complex c(2)M and c(3)G18,19 and members of the Chromosome Passenger Complex such as INCENP, and Aurora B kinase20. Genes that are required for oocyte and early embryonic development are also upregulated in the ovary, such as nanos, vasa, and bicoid21,22,23,24. By considering a gene’s expression pattern and sequence homology, it is possible to predict the function and subcellular localization of novel proteins. Such information would allow prioritization of screens based on the likelihood of a gene being involved in a biological process such as meiosis. Several online databases provide tissue-specific expression profiles and functional annotations of Drosophila genome, and they could be used to identify potential genes required for meiosis and fertility15,25,26,27. In this study, we identified 94 novel genes involved in meiosis and fertility by combining tissue-expression profile, gene annotation, functional analysis with RNAi knockdown, and cytological analysis. When these genes were knocked down in the germline, the flies displayed meiosis or germline development related phenotypes, including sterility, reduced fertility, and elevated levels of nondisjunction. These genes are excellent candidates for future mechanistic studies in flies and mammals.
Methods
Candidate gene selection from FlyAtlas 1
Gene expression profiles in different fly tissues were downloaded from FlyAtlas 1 (http://flyatlas.org/data.html)15. A total of 18,770 probe sets from 13,500 genes were included on the Affymetrix Drosophila Genome 2 expression array. The gene names and Flybase IDs were validated and corrected by FlyBase (https://flybase.org/). Genes with the following symbols in their names were removed, including unknown genes (“---”), non-protein-coding genes (“CR”), RNA genes (“rna”), and ribosomal proteins (“rps”/“rpl”). Expressions in tissue “Drosophila S2 cells” were excluded from analysis.
For each gene, the enrichment value for each tissue was calculated as the tissue-specific mean expression divided by the mean of the fly whole body; the p value was calculated using two-tailed Student’s t test. The expression direction is categorized as “up” when the enrichment value is larger than 1 and the p value is less than or equal to 0.05 (enrichment > 1 and p-value ≤ 0.05), “down” as enrichment < 1 and p-value ≤ 0.05, and “others” for anything else. To select genes with high confidence, microarray probe sets with no signals detected among four biological replicates (i.e., no “present” calls) in ovary, testis, and larvae central nervous system (CNS) were removed. One representative probe set was selected for each gene by selecting the one with the most biological replicate results and direction being “up” in ovary, testis, and larvae CNS. The selected genes were divided in four groups: ovary_only (“up” in ovary expression and not “up” in other tissues), ovary + cns (“up” in ovary and larvae CNS expression, not “up” in other tissues), ovary + testis (“up” in ovary and testis expression, not “up” in other tissues), and ovary + CNS + testis (“up” in ovary, larvae CNS, and testis expression, not “up” in other tissues).
Candidate gene annotation and prioritization for experimental validation
To identify novel meiosis gene for functional validation, gene symbols with the format “CG + numbers” were considered to be not-well studied and selected. Next, the transgeneic RNA interference line for the candidate genes were searched in the Transgenic RNAi Project (TRiP)28,29 at Bloomington Drosophila Stock Center (https://bdsc.indiana.edu/index.html). Candidate genes without TRiP stocks were removed.
Experimentally-validated and predicted meiosis genes in Drosophila were extracted from MeiosisOnline (https://mcg.ustc.edu.cn/bsc/meiosis/index.html)30. Gene expression value of three ovary cell clusters, germ cells (GC), germarium soma, and follicle cells (FC), were calculated using single-cell transcriptome data31. For each cluster, the expression level of each gene was calculated as the median value of expressions from individual stages as defined in the Supplemental Table S2 of31.
Candidate gene annotation, enrichment and protein–protein interaction network analyses
Orthologs and known alleles of candidate genes were annotated using FlyBase27. Enrichment analyses were performed for positive genes with human and mouse orthologs using the overrepresentation analysis in ConsensusPathDB (CPDB, http://cpdb.molgen.mpg.de/)32. Enriched terms (e.g., Gene Ontology, pathway, or protein complex) containing at least two input genes were selected for further analysis. Enrichment p values were determined by CPDB using a hypergeometric test and q values represent the adjusted p values using the false discovery rate method.
Protein–protein interaction (PPI) network of positive genes was constructed using interactions information provided by STRING Drosophila melanogaster database33. Interactions with at least medium confidence (i.e., combined scores ≥ 0.4) were selected.
RNAi knockdown, sterility and nondisjunction (NDJ) assays, and RT-PCR
Potential meiotic genes were screened using RNA interference (RNAi). Stocks for RNAi were obtained from the Bloomington Drosophila Stock Center at the Indiana University Bloomington (https://bdsc.indiana.edu/index.html). In these transgenes, the shRNA sequence is placed downstream of the UAS enhancer, thus requiring the presence of the GAL4 activator to begin gene knockdown via RNAi.
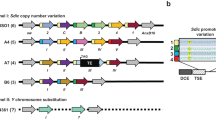
Crosses were set up using 10–20 UAS:shRNA males and 15–25 females with a tissue-specific GAL4. Each shRNA was crossed to two GAL4 stocks to induce expression (Fig. 1B). P{GAL4: :VP16-nos.UTR}CG6325MVD1 (referred to as MVD1) initiates expression of transgenes in the pre-meiotic mitotic cells of the germline and continues throughout oocyte development34. P{w[+ mC] = matalpha4-GAL-VP16}V37 (referred to as matα) expression begins late in prophase (region 2b/3), after early pachytene and the formation of crossovers and the synaptonemal complex, and continues through late prophase until stage 14 oocyte20,35.
Analysis workflow. (A) The original 13,500 genes in Flyatlas1 were first selected by different enrichment and prioritization filters. The passed genes were validated by RNAi knockdown and sterility and nondisjunction assays. A total of 94 genes and the associated 110 RNAi lines were identified as positive meiosis genes. The numbers of genes in different sub-categories (e.g., ovary_only) were counted. The numbers of genes (n(genes)) and RNAi lines (n(RNAi lines)) after each filter were denoted. (B) Schematic of the Drosophila ovary and expression patterns of P{GAL4: :VP16-nos.UTR}CG6325MVD1 (referred to as MVD1) and P{w[+ mC] = matalpha4-GAL-VP16}V37 (referred to as matα).
All crosses were kept at 25 °C and were allowed to incubate for 10 days until progeny began to emerge. Virgin female progeny from this cross that carried both the UAS RNAi and the GAL4, as indicated by the y + and w + phenotypes, were collected. Five UAS RNAi/GAL4 females were then crossed to five y w/Y, Bs males in five sets of vials (replicates). On days 14 and 18, the crosses were scored by recording the number of wildtype females (X/X), Bar males (X/BS Y), aneuploid males (X/O), and aneuploid females (X/X/BS Y). The frequency of NDJ was calculated as 2(XO + XXY)/[2(XO + XXY) + X/X + X/Y]. An RNAi line is considered positive if the cross has at least one of the following phenotypes: 1) sterile; 2) NDJ frequency ≥ 1.7% in either MVD1 or matα, or 3) ≤ 100 progeny in either MVD1 or matα. A candidate gene is considered a positive gene if at least one of its shRNA lines is positive. Some positives from these two tests were crossed to P{w[+ mC] = tubP-GAL4}LL7 (referred to as Tub-Gal4), which expresses throughout the whole body of the fly and is a test for a somatic function such as mitosis. For several candidate genes, mutations were available from either the Bloomington Drosophila Stock Center or the National Institute of Genetics Fly Stocks (the NIG-Fly sgRNA and KO collections).
For reverse transcriptase quantitative PCR (RT-qPCR), total RNA was extracted from late-stage oocytes using TRIzol® Reagent (Life Technologies). cDNA was consequently prepared using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The qPCR was performed in a StepOnePlus™ (Life Technologies) PCR system using TaqMan® Gene Expression Assays (Life Technologies).
Cytology
To study the effects of target gene knockdowns on germline development, meiosis, and embryonic development, the gross morphology of the ovaries was examined using a dissecting microscope. Small ovaries are indicative of a failure in germline development, such as loss of germline stem cells or development of the ovarian cysts. For cytological examination of early (germarium/ pachytene) or late (stage 14/Metaphase I) meiosis, oocytes were collected and examined using immunofluorescence. Crosses were set up using 10–20 UAS:shRNA males and 15–25 tissue-specific GAL4 females. Twelve days following the cross, approximately 20 (for germarium) or ~ 300 (for stage 14) progeny that carried both the UAS regulated RNAi (y+) and the tissue-specific GAL4 (w+) were collected and fed yeast for 2 days at 25 °C to promote egg laying. After 2 days, oocytes were collected and fixed using whole mounts to preserve the three-dimensional structure. Details of the two fixation protocols are described elsewhere36,37.
The primary antibodies used were: mouse anti-C(3)G (1:500)38, rabbit anti-C(2)M (1:400)18, a combination of two mouse anti-ORB antibodies (4H8 and 6H4, 1:100)39, rat anti-VASA (1:200), rabbit anti γH2AV (1:500)40, mouse anti-α-tubulin conjugated to FITC (1:50) to stain microtubules, rabbit anti-CID (1:1000), and rat anti-INCENP (1:600). Additionally, Hoechst 33342 was used to stain for DNA. Following overnight incubation, oocytes were washed and stained with secondary antibodies for 4 h at room temperature. The secondary antibodies used were: goat anti-rat Cy3 (1:100), goat anti-rabbit 647 (1:100), and Alexa 488 (1:200) (Jackson Labs and Invitrogen). Oocytes were then mounted in SlowFade Gold (Invitrogen) and imaged using a Leica TCS SP8 confocal microscope with a 63x, NA 1.4 lens. All images shown are maximum projections of complete image stacks. Statistical analysis of sister kinetochore separation and foci quantification were performed using the microscopy image analysis software Imaris (Oxford Instruments).
Results
Selection of genes up-regulated in the ovary
FlyAtlas 1 contains mRNA expression levels of 13,500 genes in Drosophila adult and larval tissues. We selected 10,948 genes that are expressed in ovary, testis, and larval CNS. Genes that were up-regulated in adult ovaries while not up-regulated in other tissues except testis and larval CNS were selected as initial genes of interest (see Methods for detail). This selection resulted in 1118 genes, 611 of which were up-regulated only in ovary (referred to as “ovary-specific” in the following text). The ovary-specific genes included known meiotic genes, such as c(2)M and c(3)G. Among the 1118 genes, 141 were up-regulated in both ovary and testis. These genes were included because testis is a meiotic tissue. Genes in this selection included ord, a gene required for sister chromatid cohesion. Finally, because of mechanistic conservation in segregating chromosomes, we included genes required for meiosis that are also required for mitosis. Therefore, we included genes up-regulated in ovary and larval CNS because the larval CNS is a mitotically active tissue. Among genes upregulated in ovary, 306 were also up-regulated in CNS and 60 were up-regulated in both CNS and testis (Fig. 1A). Examples of these genes included spindle-associated proteins such as the CPC components Incenp and aurora kinase B (aurB), and kinetochore proteins such as Ndc80 and Spc105R.
Among the 1118 genes, 974 have human orthologs and 964 have mouse orthologs (Table S1). Importantly, 873 of these genes have a shRNA TRiP stock which makes phenotype screening feasible28. To identify potential novel meiosis genes, we then selected a subset of uncharacterized genes to examine, many of which lack a gene name, and for whom a shRNA TRiP stock was available. This group was enriched for genes that have not been studied before and have no known function. After further manual review, we selected 242 genes for fertility and non-disjunction assessment after RNAi knockdown (Fig. 1A). Since beginning this study some genes that were only known by a CG name have since been named (Table 1, Table S2).
Ovary-specific RNAi knockdown of candidate genes
Each shRNA strain was crossed to one of two GAL4 expressing females to induce shRNA expression (Fig. 1B). P{GAL4: VP16-nos.UTR}CG6325MVD1 (referred to as MVD1) initiates expression of transgenes specifically in the pre-meiotic cyst cells of the germline and expression continues throughout oocyte maturation34. P{w[+ mC] = matalpha4-GAL-VP16}V37 (referred to as matα) expression begins in prophase (region2b/3) and expression continues through Metaphase I in stage 14 oocytes20,35. A significant phenotype (a “positive”) was recorded if the RNAi knockdown produced sterile females, or an increased nondisjunction (NDJ) rate (≥ 1.7%) that was higher than most of RNAi lines (Figure S1), or a small brood size (< 100). Because each gene was tested with MVD1 and/or matα promoters, and because some genes were tested using multiple shRNA lines, we defined a positive gene as one with at least one RNAi experiment yielding a positive result with either the MVD1 or the matα promoter.
Following completion of nondisjunction and sterility assays on 242 novel gene candidates in 301 RNAi lines (Table S2), we identified 94 genes that showed evidence of a function in germline development, meiosis, or embryonic development in experiments with 110 RNAi lines (Table 1, Fig. 2). Some positive RNAi lines (e.g., GL00570) were present in multiple phenotype categories (NDJ, Sterile, and small brood size) because: (1) different phenotypes were observed between the MVD1 and the matα experiments; (2) phenotypes of some shRNAs met the criteria of both NDJ and small brood size (Fig. 2A). Among the 94 genes, 57 are ovary-specific genes, 22 are expressed in both ovary and larval CNS, 12 are expressed in both ovary and testis, and 3 are expressed in all three tissues. Knockdown of 29 genes produced sterile females, 24 genes had an increased nondisjunction rate (≥ 1.7%), and 18 genes produced small brood sizes (< 100) (Fig. 2B). The remaining 23 genes showed different phenotypes in MVD1 and matα crosses and/or with different shRNAs (overlapping sections of the Venn diagram, Fig. 2B). For example, CG10336 MVD1/shRNA knockdown females were sterile, but had a small brood size in matα/shRNA females. The difference between the two GAL4 lines provides temporal information on when these genes function during oocyte development.
Venn diagrams for (A) 110 positive RNAi lines and (B) 94 positive genes. Some RNAi lines and associated positive genes are present in multiple phenotype categories (NDJ, Sterile, and small brood size (Small_brood)) because: (1) different phenotypes were observed in the MVD1 and matα experiments; (2) phenotypes of some lines met the criteria of both NDJ and small brood size. For example, RNAi line GL00570 and the associated CG13741 gene was Sterile in MVD1 and meet both NDJ and small brood size criteria in matα. In addition, CG3430 was tested with multiple RNAi lines and are present in all phenotype categories. See details in Table 1.
To confirm that the RNAi approach was effective, five positive genes that produced sterile females were tested by qRT-PCR. All showed significant knockdowns of the intended genes (Table 2, CG4951, CG18787, CG3430, CG10336, CG8142).
Annotation and enrichment analyses of positive genes
To identify genes with a role in meiosis, we examined proposed functions of each identified gene in other species (Table 1). Among the 94 genes, 86 genes have both human or mouse orthologs. Using human or mouse orthologs of the Drosophila genes, we performed gene ontology (GO) and pathway enrichment analyses (Tables S3, S4). Several GO terms showed significant enrichment in human and mouse orthologs, such as replication fork (q < 5e−5 in human and mouse, Table S3). Similarly, several pathways showed significant enrichment, such as DNA replication (q < 2.7e−4 in mouse), DNA repair (q = 0.016 in mouse, q = 0.0024 in human, Table S4). These GO terms and pathways might be expected as important for various steps of meiosis or the mitotic divisions of the early embryo. In addition, genes associated with RNA biology, such as spliceosome complex, RNA helicase activity, and ribosome biogenesis, also appeared frequently in the analysis. These findings are consistent with the known importance of regulating RNAs in oocyte and early embryonic development41,42,43.
Furthermore, we examined the interaction among the candidate genes by constructing a protein–protein interaction (PPI) network. We identified a network of 67 positive genes in one main cluster (Table S5, Fig. 3). The cluster contains genes in two significantly enriched GO terms related to cellular component biogenesis and mRNA splicing: ribonucleoprotein complex biogenesis (q < 3.1e−5 in mouse, CG32344, CG4554, CG13096, CG11188, CG11660, and CG7993) and spliceosome complex (q < 1.5e−5 in human, CG8435, CG4980, CG33228, CG11985, CG6610, CG7971, CG4849, and CG4973). These genes could be required for generating oocytes with enough maternal products to support embryonic development. We also found three genes in the pathway HDR through Homologous Recombination (HRR) or Single Strand Annealing (SSA) (CG8142, CG10336, CG12018, CG15220, and CG10981). These genes could be important for meiosis as homology-directed repair is crucial during meiosis to ensure proper chromosome segregation6.
Protein–protein interaction network of candidate genes. Only connected genes are shown. Genes present in three representative enriched GO terms: ribonucleoprotein complex biogenesis, spliceosomal complex, and HDR through homologous recombination (HRR) or single-strand annealing (SSA) are highlighted with the enriched terms labeled. See Supplemental Table S5 for a full list of interactions.
While we selected genes expressed in the ovary, ovaries are a complex tissue consisting of somatic and germline cell types. Insights into the function of these genes could come from identifying in which ovarian cell type they are expressed. A single cell ovary transcriptome dataset has identified several somatic (germarium soma, follicle cell (FC)) and germline cell (GC) cell types in ovaries31. Based on this dataset, 907 of the 1118 ovary up-regulated genes in our initial dataset (81%) had higher expression in the GC cluster compared to the germarium soma and FC clusters, with 85% of positive genes (80 genes) showing the same trend. These results suggest that our candidate genes are highly enriched in germline-specific genes and are likely to function in oocyte development.
Analysis of ovaries from select shRNA lines: gene with germline-specific phenotypes
Some genes are expected to be specific to the germline. These genes are predicted to have low expression in the central nervous system and be viable when the shRNA is expressed with whole-body promoter Tub-Gal4. Genes that fit this pattern are good candidates for germline-specific genes (Table 1). Examples of germline-specific genes include CG11133, CG18787, CG4951, CG5877 and CG8435. In some cases, the tissue gene expression data did not predict the somatic phenotype with Tub-Gal4. For example, CG10336 encodes the orthologue of human TIPIN. Although expressed in the nervous system, shRNA GLC01611 was viable when expressed with Tub-Gal4. This is the expected phenotype for genes that may not be required for ovary development unless there is an inducer of DNA damage44, which may include CG10336. In some other cases, Tub-Gal4 crosses might not provide expected results. For example, if Tub-Gal4 is provided maternally, then it could drive expression of the shRNA in embryo and cause embryonic lethality. Null alleles could survive, however, because the maternal contribution of wild-type protein allows them to survive. An example of a candidate in this class is CG13741, also known as Bootlegger (Boot). Boot is an ovary-specific gene, and null alleles are viable but female sterile45, although the shRNA expressed with Tub-Gal4 was lethal. Similarly, null alleles of CG9203 are viable but female sterile, but shRNA expressed with Tub-Gal4 was lethal. CG9203 is also known as maternal haploid (mh), a gene required for fusion of male and female pronuclei46,47. The roles of both of these genes in meiosis have not been studied.
CG18787 knockdown has premature karyosome and sister centromere separation
One shRNA targeting CG18787 could also target a second gene, CG18789. These two genes are located in close proximity to one another, separated by approximately 2.5 kb and two other genes. CG18787 encodes a 398 amino-acid protein that has 99% amino acid identity with CG18789, with only 3 amino acid differences. Given the high similarity and the relative location of these two genes within the genome, it is likely that CG18787 and CG18789 arose from a recent gene duplication event. shRNA HMC03818 produced a more severe fertility defect when crossed with both the MVD1 and matα compared to shRNA HMC04063 (Table 1). Analysis of the sequences of each shRNA demonstrated that HMC03818 could target both CG18787 and CG18789, while HMC04063 probably targets only CG18787. Thus, HMC03818 could cause a more severe fertility phenotype than HMC04063 because its shRNA targets both genes. HMC03818 did not cause lethality when crossed to Tub-Gal4, indicating CG18787 may not be required for mitosis, and consistent with its low expression in the central nervous system.
Mature oocytes expressing HMC03818 had severe karyosome separation when compared to wildtype oocytes (54%, n = 24, wildtype: Fig. 4A; two examples of HMC03818: Fig. 4C,D). This severe phenotype is consistent with a defect in sister chromatid cohesion during meiosis48,49. In addition, the separation of sister kinetochores is a phenotype commonly associated with a loss of centromere cohesion50. Therefore, we determined the frequency of sister kinetochore separation in HMC03818/ matα oocytes. A normal meiosis is expected to have 8 foci, one for each pair of sister centromeres, although usually less is observed due to clustering of the centromeres. (avg = 6.9, Fig. 4E). In contrast, HMC03818 RNAi oocytes had a significant elevated frequency of sister kinetochore separation (avg = 8.8, p = 0.0185, Fig. 4E). These results support the conclusion that G18787 is required for sister chromatid cohesion.
Cytological analysis of Drosophila oocytes. Drosophila oocytes were extracted and stained for Tubulin, the centromere (CID/CENP-A), central spindle (INCENP), and DNA (Hoechst). Scale bars are 5 μm. (A) An example of a wild-type Metaphase I arrested spindle with the formation of a singular karyosome and symmetric division of the centromeres. Knockdown using matα of (B) CG4951 and (C,D) CG18787. Inset in each panel shows the karyosome (DNA). (E) Quantification of sister kinetochore foci for both wildtype and CG18787 RNAi oocytes.
The cohesion defects were observed using HMC03818/ matα, and because meiotic cohesion is probably established during S-phase before matα expression48, these results suggest G18787 has a cohesion maintenance function. In addition, mutants defective in establishing sister chromatid cohesion also have defects in Synaptonemal Complex (SC) assembly51,52. We examined early meiotic prophase oocytes from HMC03818/MVD1 females and found no defects in SC component C(3)G and double-strand break (DSB) marker γH2AV (wildtype: Fig. 5A; HMC03818 / MVD1: Fig. 5B). These results suggest that the defects associated with the loss of CG18787 and CG18789 may affect the maintenance of cohesion in oocytes. Mature oocytes were absent in HMC03818 / MVD1 ovaries (Table 1), suggesting that CG18787 is also required for the mitotic divisions of germline development.
Early prophase (germarium) images. Ovaries were dissected from shRNA / MVD1 females and stained for C(3)G (SC, green), γH2AV (DSBs, red), CENP-C (centromeres, white), and DNA (blue). Panels: wild-type (A), RNAi against CG18187 (B), CG3430 (C), RPA3 (D), and CG12259 (E). The germarium is divided into four stages: 1 (mitotic region with stems cells, and 2, 4, and 8 cell cysts), 2 (16 cell cysts in early meiosis, including zygotene and pachytene), and 2b and 3 (mid pachytene 16 cell cysts). The ovaries in C-E lack 16 cell cysts. Some of the cells are somatic follicle cells and lack meiotic markers like C(3)G. The scale bars are 5 µm.
CG18787 contains a nucleoporin domain and has limited homology to human NUP42. This is significant because recent studies have found that upon nuclear envelope breakdown leading up to cellular division, many nucleoporins localize to kinetochores and serve alternative functions to aid in cellular division. An example of this is the nucleoporin Elys, which was found to function as a Protein Phosphatase 1 scaffold during M-phase exit and thus aids in the disassembly of kinetochores53. Based on these results, we believe that CG18787 has a meiotic function required for the maintenance of sister chromatid cohesion.
CG4951 may be required for biorientation
Knockdown of CG4951 using shRNA GL01154 caused sterility (Table 1). GL01154 did not cause lethality when crossed to Tub-Gal4, suggesting the function of CG4951 is germline-specific and not required for mitosis. This phenotype is consistent with its expression pattern, which shows low expression in the nervous system. Sequence analysis of this gene does not provide any significant insight into the function as the predicted protein sequence does not contain identifiable protein domains or regions of high conservation. CG4951 is an example of a poorly conserved gene. It encodes a 320 amino-acid protein that is conserved within the Drosophila genus but not found in other Diptera such as the mosquito. Cytological analysis of CG4951 RNAi oocytes did not reveal significant defects in spindle assembly (Fig. 4B). Further studies are needed to determine if there are biorientation defects, as we observed asymmetric distribution of centromeres, which would result in the abnormal division of the chromosomes at anaphase I (Fig. 4B, 2/10 oocytes). This phenotype would suggest that CG4951 is involved in the regulation of kinetochore biorientation.
Genes with somatic phenotypes
Genes expressed at high levels in the nervous system could have somatic phenotypes. Lethality in the Tub-Gal4 experiment, however, identifies any gene with a function in somatic tissue (Table 1). For example, CG16838 is enhanced level of genomic instability 1 (elg1). We have confirmed that most homozygous mutants (elg1/elg2) are lethal, with some rare but sick survivors, consistent with previous studies54. Consistent with this result, the shRNA expressed with Tub-Gal4 caused lethality, although there were a few sick survivors. Interestingly, we identified another gene that probably interacts with elg1, CG8142, also known as RFC4 in human. This gene is also expressed at high levels in the nervous system and shRNA GL00569 is lethal with Tub-Gal4. One possibility, based on sequence homology, is that these genes are required for DNA replication in the germline. This would explain the lack of oocytes with shRNA for CG8142 expressed with MVD1. However, elg1 shRNA expressed with matα also failed to make oocytes, which cannot be a DNA replication defect because there is none in maturing oocytes. Further work is required to understand the defects, similarities, and differences, between these two genes.
Additional examples of genes required in the germline and somatic cells include CG15220, also known as RPA3, which failed to make oocytes when depleted with MVD1, was sterile with matα, and was lethal with Tub-Gal4. CG7185 is known as Cleavage and polyadenylation specific factor 6 (Cpsf6) and is an essential gene55, consistent with its high expression in ovaries and the larval nervous system. CG7033 (Chaperonin containing TCP1 subunit 2 (CCT2)) is also a gene in this category but had a unique phenotype with Tub-Gal4. The crosses were sterile, indicating the F1 embryos died. A similar phenotype was observed with CG33217, which is orthologous to human PELP1 (proline, glutamate and leucine rich protein 1). Using shRNA GL01509, the F1 progeny from a cross to Tub-Gal4 were lethal. These genes have an unusual dosage sensitivity as this must represent expression induced by maternal contribution of Tub-Gal4.
CG3430 knockdown has biorientation defects and precocious anaphase I onset
Two shRNAs were associated with CG3430 (Table 1). The first shRNA, GL01184, caused reduced fertility and a loss of developing oocytes (see details below) when expressed with MVD1, and was sterile when expressed with matα. The second shRNA, HMC06551, had little effect when expressed with MVD1 and elevated levels of nondisjunction (15.9%) when expressed with matα. The differences in these phenotypes indicate that GL01184 resulted in a stronger knockdown of mRNA than HMC06551. Both shRNAs caused lethality when expressed by Tub-Gal4, indicating this gene could be required for mitosis as well. Furthermore, two CRISPR alleles were obtained from NIG-FLY, and were lethal as trans heterozygotes (CG3430SK5/CG3430SK7).
By sequence comparison, we found that CG3430 is homologous to the Mini-Chromosome Maintenance Complex Binding Protein (MCMBP) superfamily. For example, CG3430 has 37% identity with human MCMBP, which was identified as a protein that associates with and promotes assembly of the MCM2-7 complex56,57. Other functions have also been suggested. In Xenopus, MCMBP promotes disassembly of MCM complexes from chromatin58. Arabidopsis thaliana MCMBP/ETG1 appears to be needed for sister chromatid cohesion59. Our results with matα demonstrate that Drosophila MCMBP has a function after premeiotic S-phase in oocytes.
Genes required for early germline development
As noted above, shRNA targeting CG8142 or CG3430 expressed with MVD1 resulted in ovaries lacking developing oocytes. To identify defects early in germline development when CG3430 was depleted, ovaries from GL01184/MVD1 females were dissected and stained with antibodies that mark SC component C(3)G and the DSB marker γH2AV. We observed a defect early in germline development. The ovaries failed to make 16 cell cysts, and oocytes with full SC formation and γH2AV foci were not observed (Fig. 5C). Thus, CG3430 is required prior to meiosis, for the stem cell divisions or the mitotic divisions that generate the 16 cell cysts. Similarly, CG15220, encoding the Drosophila orthologue of Replication protein 3A (RPA3), is required for germline development, as shown by the absence of oocytes with full SC formation and the DSB marker γH2AV in HMJ24068/MVD1 ovaries (Fig. 5D).
We observed a similar early germline defect with several other genes. Three lines of evidence suggest that CG12259 is only required early in germ line development. First, HMJ23711/MVD1 females were sterile, but HMJ23711/matα females were fertile. Second, cytological analysis of HMJ23711/MVD1 females revealed a severe phenotype, with a defect early in germline development, and failing to make 16 cell cysts with an oocyte containing full SC formation or the DSB marker γH2AV (Fig. 5E). We examined these ovaries with VASA staining, which is a marker specific for germline cells. Compared to wild-type ovaries, HMJ23711/ MVD1 ovaries had a deficiency in cells with cytoplasmic VASA, indicating a loss of germline cells (wildtype: Fig. 6A; HMJ23711/MVD1: Fig. 6B). Thus, this gene may be required prior to meiosis, during the mitotic divisions that generate the 16 cell cysts. Indeed, the loss of germ cells in HMJ23711/MVD1 ovaries is consistent with a defect in maintaining the stem cell population of the ovary. Third, HMJ23711 did not cause lethality when crossed to Tub-Gal4, indicating CG12259 may not be required for mitosis and is germline specific. CG12259 has the highest homology with FAM50A in mammals with possible RNA/nucleic acid binding activity and functions in chromatin organization60.
Whole germarium images. Ovaries were dissected from shRNA / MVD1 females and stained for C(3)G (SC, green), VASA, (germline cells, red), CENP-C (centromere, white), and DNA (blue). Panels: wild-type (A), RNAi against (B) CG1229 and (C) Odj. B and C have relatively small ovaries characterized by a lack of 16-cell cyst formation and C(3)G limited to the centromeres, indicating a loss of germ cells. The cells lacking VASA staining in A-C are somatic cells. The scale bars are 5 µm.
Two shRNAs for CG7357 (HMC05191 and HMJ23468), also known as Oddjob (Odj), had a similar phenotype in MVD1 ovaries. Oddjob is a member of the ZAD family of zinc-finger transcription factors. There are no known mutations, although a similar result was observed using the same RNAi lines in a screen of ZAD transcription factors for functions in the female germline61. This function appears to be specific for the early germline because, like CG12259, shRNA/matα females were fertile. Also, like CG12259, shRNA/ MVD1 ovaries lacked germ cells as shown by VASA staining, indicating Oddjob is required for maintaining the germline, possibly the stem cell population (Fig. 6C). However, Oddjob also has a somatic function, as suggested by its high expression in the nervous system and lethality of HMJ23468 with Tub-Gal4.
Conclusion
Sexual reproduction depends on two integrated processes, the faithful transmission of chromosomes during meiosis to yield viable gametes, and the development of a gamete capable of fertilization and supporting embryonic development. Our unbiased screen in Drosophila has identified candidate genes that could be further studied to expand our understanding of these critical processes.
In this study, we leveraged gene expression profiles and functional annotation to identify novel meiotic genes followed by phenotyping validation in Drosophila RNAi lines. We identified 94 genes that displayed elevated levels of nondisjunction or loss of fertility when knocked down, showing that these genes are required in multiple phases of gametogenesis and meiosis. We screened 301 RNAi lines using two GAL4 promoters, which resulted in a selection for genes required early in gametogenesis or late in oogenesis. Only one of our 94 meiosis candidate genes (CG42307///mus312) were found in the manually curated meiosis database, MeiosisOnline30. This is probably because, by design, most of the genes we tested were not previously characterized. These results support our approach for identifying novel meiosis and fertility genes.
At least two previous studies used RNAi to screen for genes related to the loss of fertility. First, was a screen using a GAL4 promoter with similar expression characteristics to MVD1 and matα for maternal proteins that are phospho-regulated62. Among the 132 genes whose knockdown affected oogenesis (class 2 to class 6), three genes overlap our 1118 ovary up-regulated genes, including one of our positive genes CG11188 and two genes that we tested but showed a normal phenotype (CG6961, CG4968). The most likely reason for lack of overlap is the different selection criteria, as focusing on a protein modification (phospho-regulated) may select for many genes that are not ovary enriched. Second was a screen using a GAL4 similar to MVD1 for genes required for germline stem cell maintenance63. Among the 366 positive genes, 10 overlaps with our positive results (CG13096, CG8435, CG7033, CG8728, CG7185, CG8116, CG11985, CG11660, CG42307 and CG30020) and two overlaps with our negative results (CG9548 and CG11398). The lack of overlap in this case is more surprising, but could be due to the screening phenotype (stem cell maintenance) compared to defects in fertility, or differences in how the genes for screening were selected.
The lack of overlap of these previous studies with our positive genes substantiates the validity of our approach of identifying novel genes. Future studies could continue novel gene discovery by integrating ovary up-regulated genes in FlyAtlas 1 and FlyAtlas 225. Although there are differences in experimental design and data generation methods between the two databases, the majority of the FlyAtlas1 ovary up-regulated genes (72.7%) are also up-regulated in FlyAtlas2, including 66% of the positive genes. Another future direction would be to test these genes for a role in the male germline, using appropriate Gal4 activators such as MVD1 and bam-GAL4.VP1664.
The genes we discovered fall into a variety of functional classes. The use of the two GAL4 lines to induce expression of the shRNA also provides useful temporal information. Some genes, such as CG15863, CG5877, CG30020, CG42232, and CG7357, were sterile with MVD1 but not matα, indicating they had a function only in early germline development. Conversely, genes such as CG17361, CG6951, CG6540, and CG4554 were sterile with matα but not MVD1, suggesting they only function late in oocyte development. A surprising number of genes produced no mature oocytes with matα. This phenotype with matα, such as CG3430, CG14174, CG16838, CG33217, and CG6540, suggests a role in growth of the oocyte during the meiotic prophase arrest. We also identified several genes with a reduced fertility phenotype. This could indicate a partial mRNA knockdown or a function important but not essential for fertility.
Several genes, such as CG6937, CG3407, CG3430, CG12259, CG10635, CG10344, CG6843, and CG34261, had significant nondisjunction phenotypes and thus appear to be required for meiosis. Although several genes involved in homolog pairing and recombination within Drosophila show little apparent sequence homology, there is strong genetic and structural conservation of meiosis across eukaryotes65,66,67,68. The functional similarity between human and fruit fly establishes the foundation that understanding of the Drosophila genome could provide a valuable source to yield insights into human gene functions that are not easily obtainable in mammals like humans or mice. For example, the positive gene CG8915 is the homolog of a potential human meiotic gene YTHDC2 (mouse Ythdc2), which plays a role in regulating meiosis in human and mouse69,70,71.
An important outcome of this screen is the identification of genes with no known ovary function that have homologs in higher eukaryotic systems. For example, 86 of the 94 genes have mouse homologs, and about one-third of these genes have no known function in meiosis or reproduction likely because they have not yet been examined. The high percentage of human/mouse homologs in our positive genes demonstrate this approach enriches for genes that are crucial for oocyte and embryo development and can uncover novel mechanisms for female infertility. This screen also uncovered nearly 20 genes that either function or have predicted function in RNA biology, a function intimately linked to high egg quality. Therefore, this study has opened new and critical areas related to meiosis and egg quality that should be explored.
Data availability
The data is available in the manuscript and the associated Supplemental Tables.
References
Nagaoka, S. I., Hassold, T. J. & Hunt, P. A. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504. https://doi.org/10.1038/nrg3245 (2012).
Hassold, T. & Chiu, D. Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum. Genet. 70, 11–17 (1985).
Biswas, L. et al. Meiosis interrupted: The genetics of female infertility via meiotic failure. Reproduction 161, R13–R35. https://doi.org/10.1530/REP-20-0422 (2021).
Volozonoka, L. et al. A systematic review and standardized clinical validity assessment of genes involved in female reproductive failure. Reproduction 163, 351–363. https://doi.org/10.1530/REP-21-0486 (2022).
Capalbo, A. et al. Preconception genome medicine: current state and future perspectives to improve infertility diagnosis and reproductive and health outcomes based on individual genomic data. Hum. Reprod. Update 27, 254–279. https://doi.org/10.1093/humupd/dmaa044 (2021).
Hughes, S. E., Miller, D. E., Miller, A. L. & Hawley, R. S. Female meiosis: Synapsis, recombination, and segregation in Drosophila melanogaster. Genetics 208, 875–908. https://doi.org/10.1534/genetics.117.300081 (2018).
Hinnant, T. D., Merkle, J. A. & Ables, E. T. Coordinating proliferation, polarity, and cell fate in the Drosophila female germline. Front. Cell Dev. Biol. 8, 19. https://doi.org/10.3389/fcell.2020.00019 (2020).
Von Stetina, J. R. & Orr-Weaver, T. L. Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb. Perspect. Biol. 3, a005553 (2011).
Hamm, D. C. & Harrison, M. M. Regulatory principles governing the maternal-to-zygotic transition: Insights from Drosophila melanogaster. Open Biol. 8, 180183. https://doi.org/10.1098/rsob.180183 (2018).
Krauchunas, A. R. & Wolfner, M. F. Molecular changes during egg activation. Curr. Top. Dev. Biol. 102, 267–292. https://doi.org/10.1016/b978-0-12-416024-8.00010-6 (2013).
Avilés-Pagán, E. E. & Orr-Weaver, T. L. Activating embryonic development in Drosophila. Semin. Cell Dev. Biol. 84, 100–110. https://doi.org/10.1016/j.semcdb.2018.02.019 (2018).
Ding, X. & Schimenti, J. C. Strategies to identify genetic variants causing infertility. Trends Mol. Med. 27, 792–806. https://doi.org/10.1016/j.molmed.2020.12.008 (2021).
Adams, M. D. & Sekelsky, J. J. From sequence to phenotype: Reverse genetics in Drosophila melanogaster. Nat. Rev. Genet. 3, 189–198. https://doi.org/10.1038/nrg752 (2002).
Hudson, A. M. & Cooley, L. Methods for studying oogenesis. Methods 68, 207–217. https://doi.org/10.1016/j.ymeth.2014.01.005 (2014).
Chintapalli, V. R., Wang, J. & Dow, J. A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720. https://doi.org/10.1038/ng2049 (2007).
Link, N. & Bellen, H. J. Using Drosophila to drive the diagnosis and understand the mechanisms of rare human diseases. Development https://doi.org/10.1242/dev.191411 (2020).
Kaya-Çopur, A. & Schnorrer, F. A guide to genome-wide in vivo RNAi applications in Drosophila. In Methods in Molecular Biology Vol. 1478 (ed. Dahmann, C.) 117–143 (Humana Press, 2016). https://doi.org/10.1007/978-1-4939-6371-3_6.
Manheim, E. A. & McKim, K. S. The synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr. Biol. 13, 276–285 (2003).
Anderson, L. K. et al. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. U. S. A. 102, 4482–4487 (2005).
Radford, S. J., Jang, J. K. & McKim, K. S. The chromosomal passenger complex is required for meiotic acentrosomal spindle assembly and chromosome biorientation. Genetics 192, 417–429. https://doi.org/10.1534/genetics.112.143495 (2012).
Kimble, J. & Nüsslein-Volhard, C. The great small organisms of developmental genetics: Caenorhabditis elegans and Drosophila melanogaster. Dev. Biol. 485, 93–122. https://doi.org/10.1016/j.ydbio.2022.02.013 (2022).
Milas, A. & Telley, I. A. Polarity events in the Drosophila melanogaster oocyte. Front. Cell Dev. Biol. 10, 895876. https://doi.org/10.3389/fcell.2022.895876 (2022).
Schüpbach, T. Genetic screens to analyze pattern formation of egg and embryo in Drosophila: A personal history. Annu. Rev. Genet. 53, 1–18. https://doi.org/10.1146/annurev-genet-112618-043708 (2019).
Nelson, J. O., Chen, C. & Yamashita, Y. M. Germline stem cell homeostasis. Curr. Top. Dev. Biol. 135, 203–244. https://doi.org/10.1016/bs.ctdb.2019.04.006 (2019).
Leader, D. P., Krause, S. A., Pandit, A., Davies, S. A. & Dow, J. A. T. FlyAtlas 2: A new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 46, D809–D815. https://doi.org/10.1093/nar/gkx976 (2018).
The modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Gramates, L. S. et al. FlyBase: A guided tour of highlighted features. Genetics 220, iyac035. https://doi.org/10.1093/genetics/iyac035 (2022).
Ni, J. Q. et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405–407 (2011).
Perkins, L. A. et al. The transgenic RNAi Project at Harvard Medical School: Resources and validation. Genetics 201, 843–852. https://doi.org/10.1534/genetics.115.180208 (2015).
Jiang, X. et al. MeiosisOnline: A manually curated database for tracking and predicting genes associated with meiosis. Front. Cell Dev. Biol. 9, 2102 (2021).
Slaidina, M., Gupta, S., Banisch, T. U. & Lehmann, R. A single-cell atlas reveals unanticipated cell type complexity in Drosophila ovaries. Genome Res. 31, 1938–1951. https://doi.org/10.1101/gr.274340.120 (2021).
Herwig, R., Hardt, C., Lienhard, M. & Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 11, 1889–1907. https://doi.org/10.1038/nprot.2016.117 (2016).
Szklarczyk, D. et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. https://doi.org/10.1093/nar/gkw937 (2017).
Rorth, P. Gal4 in the Drosophila female germline. Mech. Dev. 78, 113–118 (1998).
Sugimura, I. & Lilly, M. A. Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev. Cell 10, 127–135 (2006).
Radford, S. J. & McKim, K. S. Techniques for imaging prometaphase and metaphase of meiosis I in fixed Drosophila oocytes. J. Vis. Exp. 116, e54666. https://doi.org/10.3791/54666 (2016).
McKim, K. S., Joyce, E. F. & Jang, J. K. Cytological analysis of meiosis in fixed Drosophila ovaries. In Methods in Molecular Biology Vol. 558 (ed. Keeney, S.) 197–216 (Humana Press, 2009).
Page, S. L. & Hawley, R. S. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15, 3130–3143 (2001).
Lantz, V., Chang, J. S., Horabin, J. I., Bopp, D. & Schedl, P. The Drosophila ORB RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8, 598–613 (1994).
Mehrotra, S., Hawley, R. S. & McKim, K. S. in Recombination and Meiosis, Crossing-Over and Disjunction Genome Dynamics and Stabitity (eds Egel, R. & Lankenau, D.) 125–152 (Springer-Verlag, 2007).
Lasko, P. Patterning the Drosophila embryo: A paradigm for RNA-based developmental genetic regulation. Wiley Interdiscip. Rev. RNA 11, e1610. https://doi.org/10.1002/wrna.1610 (2020).
Mukherjee, N. & Mukherjee, C. Germ cell ribonucleoprotein granules in different clades of life: From insects to mammals. Wiley Interdiscip. Rev. RNA 12, e1642. https://doi.org/10.1002/wrna.1642 (2021).
Weil, T. T. mRNA localization in the Drosophila germline. RNA Biol. 11, 1010–1018. https://doi.org/10.4161/rna.36097 (2014).
Sekelsky, J. DNA repair in Drosophila: Mutagens, models, and missing genes. Genetics 205, 471–490. https://doi.org/10.1534/genetics.116.186759 (2017).
Kneuss, E. et al. Specialization of the Drosophila nuclear export family protein Nxf3 for piRNA precursor export. Genes Dev. 33, 1208–1220. https://doi.org/10.1101/gad.328690.119 (2019).
Delabaere, L. et al. The Spartan ortholog maternal haploid is required for paternal chromosome integrity in the Drosophila zygote. Curr. Biol. 24, 2281–2287. https://doi.org/10.1016/j.cub.2014.08.010 (2014).
Tang, X. et al. Maternal haploid, a metalloprotease enriched at the largest satellite repeat and essential for genome integrity in Drosophila embryos. Genetics 206, 1829–1839. https://doi.org/10.1534/genetics.117.200949 (2017).
Gyuricza, M. R. et al. Dynamic and stable cohesins regulate synaptonemal complex assembly and chromosome segregation. Curr. Biol. 26, 1688–1698. https://doi.org/10.1016/j.cub.2016.05.006 (2016).
Jang, J. K. et al. Multiple pools of PP2A regulate spindle assembly, kinetochore attachments, and cohesion in Drosophila oocytes. J. Cell Sci. https://doi.org/10.1242/jcs.254037 (2021).
Wang, L. I., Das, A. & McKim, K. S. Sister centromere fusion during meiosis I depends on maintaining cohesins and destabilizing microtubule attachments. PLoS Genet. 15, e1008072. https://doi.org/10.1371/journal.pgen.1008072 (2019).
Khetani, R. S. & Bickel, S. E. Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J. Cell Sci. 120, 3123–3137 (2007).
Krishnan, B. et al. Sisters unbound is required for meiotic centromeric cohesion in Drosophila melanogaster. Genetics https://doi.org/10.1534/genetics.114.166009 (2014).
Hattersley, N. & Desai, A. The nucleoporin MEL-28/ELYS: A PP1 scaffold during M-phase exit. Cell Cycle 16, 489–490. https://doi.org/10.1080/15384101.2017.1278929 (2017).
Hayashi, R. et al. A genetic screen based on in vivo RNA imaging reveals centrosome-independent mechanisms for localizing gurken transcripts in Drosophila. G3 4, 749–760. https://doi.org/10.1534/g3.114.010462 (2014).
Tang, H. W. et al. The TORC1-regulated CPA complex rewires an RNA processing network to drive autophagy and metabolic reprogramming. Cell Metab. 27, 1040-1054.e1048. https://doi.org/10.1016/j.cmet.2018.02.023 (2018).
Sakwe, A. M., Nguyen, T., Athanasopoulos, V., Shire, K. & Frappier, L. Identification and characterization of a novel component of the human minichromosome maintenance complex. Mol. Cell. Biol. 27, 3044–3055. https://doi.org/10.1128/MCB.02384-06 (2007).
Saito, Y., Santosa, V., Ishiguro, K. I. & Kanemaki, M. T. MCMBP promotes the assembly of the MCM2–7 hetero-hexamer to ensure robust DNA replication in human cells. eLife 11, e77393. https://doi.org/10.7554/eLife.77393 (2022).
Nishiyama, A., Frappier, L. & Mechali, M. MCM-BP regulates unloading of the MCM2-7 helicase in late S phase. Genes Dev. 25, 165–175. https://doi.org/10.1101/gad.614411 (2011).
Takahashi, N. et al. The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet. 6, e1000817. https://doi.org/10.1371/journal.pgen.1000817 (2010).
Mazzarella, R., Pengue, G., Yoon, J., Jones, J. & Schlessinger, D. Differential expression of XAP5, a candidate disease gene. Genomics 45, 216–219. https://doi.org/10.1006/geno.1997.4912 (1997).
Shapiro-Kulnane, L., Bautista, O. & Salz, H. K. An RNA-interference screen in Drosophila to identify ZAD-containing C2H2 zinc finger genes that function in female germ cells. G3 https://doi.org/10.1093/g3journal/jkaa016 (2021).
Zhang, Z., Krauchunas, A. R., Huang, S. & Wolfner, M. F. Maternal proteins that are phosphoregulated upon egg activation include crucial factors for oogenesis, egg activation and embryogenesis in Drosophila melanogaster. G3 8, 3005–3018. https://doi.org/10.1534/g3.118.200578 (2018).
Yan, D. et al. A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell 28, 459–473. https://doi.org/10.1016/j.devcel.2014.01.020 (2014).
Chen, D. & McKearin, D. M. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130, 1159–1170. https://doi.org/10.1242/dev.00325 (2003).
Hemmer, L. W. & Blumenstiel, J. P. Holding it together: Rapid evolution and positive selection in the synaptonemal complex of Drosophila. BMC Evol. Biol. 16, 91. https://doi.org/10.1186/s12862-016-0670-8 (2016).
Dapper, A. L. & Payseur, B. A. Molecular evolution of the meiotic recombination pathway in mammals. Evol. Int. J. Org. Evol. 73, 2368–2389. https://doi.org/10.1111/evo.13850 (2019).
Speijer, D., Lukeš, J. & Eliáš, M. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc. Natl. Acad. Sci. U. S. A. 112, 8827–8834. https://doi.org/10.1073/pnas.1501725112 (2015).
Ramesh, M. A., Malik, S.-B. & Logsdon, J. M. A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 15, 185–191. https://doi.org/10.1016/j.cub.2005.01.003 (2005).
Conti, M. & Franciosi, F. Acquisition of oocyte competence to develop as an embryo: Integrated nuclear and cytoplasmic events. Hum. Reprod. Update 24, 245–266. https://doi.org/10.1093/humupd/dmx040 (2018).
McGlacken-Byrne, S. M. et al. Pathogenic variants in the human m6A reader YTHDC2 are associated with primary ovarian insufficiency. JCI Insight https://doi.org/10.1172/jci.insight.154671 (2022).
Saito, Y. et al. YTHDC2 control of gametogenesis requires helicase activity but not m(6)A binding. Genes Dev. 36, 180–194. https://doi.org/10.1101/gad.349190.121 (2022).
Acknowledgements
We thank Marina Druzhinina for technical assistance. Some antibodies were obtained from the Developmental Studies Hybridoma Bank, University of Iowa. Drosophila stocks were obtained from the Bloomington Drosophila Stock Center. This work is supported by NIH grants R01HD091331 and R01GM101955.
Author information
Authors and Affiliations
Contributions
Conceptualization: K.S., K.S.M., J.X.; Investigation: S.S., T.D., A.B., J.S., F.V., D.S., M.A., M.H., S.W., N.C., J.J.; Formal analysis: S.S., T.D., A.B., F.V., D.S., J.J., K.S.M., J.X.; Initial draft: S.S., K.S.M., J.X.; Supervision: K.S.M., J.X.; All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, S., Defosse, T., Boyd, A. et al. Whole transcriptome screening for novel genes involved in meiosis and fertility in Drosophila melanogaster. Sci Rep 14, 3602 (2024). https://doi.org/10.1038/s41598-024-53346-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-53346-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.