Abstract
Cyanophycin is a bacterial biopolymer used for storage of fixed nitrogen. It is composed of a backbone of L-aspartate residues with L-arginines attached to each of their side chains. Cyanophycin is produced by cyanophycin synthetase 1 (CphA1) using Arg, Asp and ATP, and is degraded in two steps. First, cyanophycinase breaks down the backbone peptide bonds, releasing β-Asp-Arg dipeptides. Then, these dipeptides are broken down into free Asp and Arg by enzymes with isoaspartyl dipeptidase activity. Two bacterial enzymes are known to possess promiscuous isoaspartyl dipeptidase activity: isoaspartyl dipeptidase (IadA) and isoaspartyl aminopeptidase (IaaA). We performed a bioinformatic analysis to investigate whether genes for cyanophycin metabolism enzymes cluster together or are spread around the microbial genomes. Many genomes showed incomplete contingents of known cyanophycin metabolizing genes, with different patterns in various bacterial clades. Cyanophycin synthetase and cyanophycinase are usually clustered together when recognizable genes for each are found within a genome. Cyanophycinase and isoaspartyl dipeptidase genes typically cluster within genomes lacking cphA1. About one-third of genomes with genes for CphA1, cyanophycinase and IaaA show these genes clustered together, while the proportion is around one-sixth for CphA1, cyanophycinase and IadA. We used X-ray crystallography and biochemical studies to characterize an IadA and an IaaA from two such clusters, in Leucothrix mucor and Roseivivax halodurans, respectively. The enzymes retained their promiscuous nature, showing that being associated with cyanophycin-related genes did not make them specific for β-Asp-Arg dipeptides derived from cyanophycin degradation.
Similar content being viewed by others
Introduction
Cyanophycin is a biopolymer first described over 100 years ago as large, light scattering granules observed in cyanobacterial cells1. These granules are composed of chains with backbones of L-aspartate residues with L-arginine attached to each Asp side chain2 (Fig. 1a). Cyanophycin contains 26% nitrogen content by mass, which, along with its inert nature and low solubility, makes it useful for nitrogen, carbon and energy storage3,4,5. Cyanophycin can be produced by a wide variety of bacteria6,7, but research in a biological context has mostly focused on cyanobacteria8,9,10,11,12. Cyanophycin is known to be especially useful for nitrogen-fixing cyanobacteria, which separate anaerobic nitrogen fixing from oxygen-producing photosynthesis either spatially in different cell types8 or temporally in a day/night cycle13.
The structure and degradation of cyanophycin. (a) Long polymer chains (typically n = 80–400) are degraded by cyanophycinase into β-Asp-Arg dipeptides, which are then hydrolyzed by isoaspartyl dipeptidases, resulting in free Asp and Arg. (b) The general reaction is catalyzed by isoaspartyl dipeptidases. X-NH = any amino acid residue.
Cyanophycin is made by cyanophycin synthetase 1 (CphA1)14 or 2 (CphA2)15 (Supplementary Fig. S1). CphA1 is a widespread enzyme that catalyzes two ATP-dependent reactions14,16: it first adds Asp to the polymer backbone and then attaches Arg to the side chain of that Asp residue through an isopeptide bond6. Some CphA1 enzymes can also incorporate lysine into cyanophycin in place of arginine, though at lower efficiency17. CphA2, a cyanobacterial enzyme related to CphA1, uses a single active site to catalyze the ATP-dependent repolymerization of β-Asp-Arg dipeptides into cyanophycin15,18.
To access the nitrogen, carbon and energy stored in cyanophycin8,13, bacteria degrade it into free amino acids. This is done in two steps (Fig. 1a, Supplementary Fig. S1): First, cyanophycin is hydrolyzed into β-Asp-Arg dipeptides by a specialized exo-cyanophycinase enzyme, either the intracellular CphB19 or CphI7, or the extracellular CphE19. Then the β-Asp-Arg dipeptides are hydrolyzed into Asp and Arg by enzymes that possess isoaspartyl-dipeptidase activity20 (Fig. 1b). The two degradation steps occur within the same cells in cyanobacterial species that have day/night regulation of cyanophycin metabolism21, while in cyanobacterial communities with cyanophycin-synthesizing heterocysts, dipeptides can be shuttled to vegetative cells for hydrolysis8. Many bacterial communities capable of using exogenous cyanophycin as a carbon and nitrogen source have been identified22,23. These communities can be found in a variety of environments, such as animal gut flora24, soil25 and fresh-water sediments26, suggesting cyanophycin is commonly found in these environments. There is evidence that the two steps of cyanophycin degradation are sometimes split between members of a bacterial consortium, where some members express cyanophycinase and others degrade the β-aspartyl dipeptides22.
Enzymes capable of degrading β-aspartyl dipeptides are very common, because β-aspartyl residues can form spontaneously from intramolecular rearrangement of Asp or Asn residues in proteins27,28. The resulting β-aspartyl dipeptides, if not degraded, can accumulate to pathological levels in cells29. In bacteria, these β-aspartyl residues can either be repaired by L-isoaspartyl O-methyltransferase enzymes (E.C 2.1.1.77)30 or be hydrolyzed into their amino acid constituents31. Two bacterial enzymes are known to have significant β-aspartyl dipeptidase activity: isoaspartyl dipeptidase (IadA)27,32, a bacterial zinc metallopeptidase; and isoaspartyl aminopeptidase (IaaA, also called plant-type asparaginase, EcAIII and IadC)20,33,34,35, a common Ntn-family enzyme with known plant and animal homologs. IadA and IaaA are evolutionarily unrelated and have different catalytic mechanisms, but both have broad substrate specificity because damage to proteins can lead to the attachment of different amino acids to Asp/Asn side chains20,32,36. Accordingly, they are also capable of degrading β-Asp-Arg/Lys, so it is assumed that β-Asp-Arg/Lys dipeptides derived from cyanophycin are degraded by general isoaspartyl dipeptidases7,19,20,37. In addition, several other enzymes, such as glycosylasparaginases, catalyze similar reactions and can display low levels of β-aspartyl dipeptidase activity38.
In this study, we analyzed the genomes in the NCBI RefSeq database39 to investigate the tendency of cyanophycin metabolism genes to co-occur and cluster together in the genome. We observe moderate levels of co-occurrence of cphA1, cyanophycinase and an isoaspartyl dipeptidase genes within these genomes. The rates of clustering of various combinations of the genes are well above random, ranging from moderate (e.g., 37 of 231 genomes containing cphA1, a cyanophycinase gene and iadA show all three genes to cluster) to high (e.g. 30 of 32 genomes with a cyanophycinase gene and iaaA, but without cphA1 genes show clustering). Characterization of the activity and structures of representative enzymes which cluster with cyanophycin synthetase and cyanophycinase genes, Leucothrix mucor IadA and Roseivivax halodurans IaaA, revealed that they have not become specific for β-Asp-Arg dipeptides.
Results
Analysis of co-occurrence of cyanophycin-metabolizing genes
To begin to quantify the occurrence, co-occurrence and clustering of cyanophycin-metabolizing genes, we first searched for the presence of cphA1, cyanophycinase (cphB, cphI or cphE) and isoaspartyl dipeptidase (iaaA20 or iadA27) in all 27,349 non-redundant, complete bacterial genomes in the NCBI RefSeq database39 at the time of analysis (Table S2). Simple occurrence data shows that isoaspartyl dipeptidases are common (found in 11,814 genomes, 43.2%), which is expected, as they have roles other than cyanophycin metabolism; that cyanophycin synthetase 1 is found in 1,614 genomes (6%); and that a recognizable cyanophycinase gene is present in 739 genomes (3%).
Co-occurrence analyses for these genes in bacteria from all clades show moderate overall rates. A recognizable cyanophycinase is present in 658 of 1,614 CphA1-encoding genomes (Table 1). Genes for IaaA or IadA are found in 1,181 of 1,614 cphA1-containing genomes, with 968 of those genomes having iaaA, 232 having iadA, and 19 having both iaaA and iadA (Table 1).
Subdividing data by bacterial clade shows distinct taxon-specific patterns (Table 2). In agreement with Flores and coworkers35, almost all complete cyanobacteria genomes in the database which encode CphA1, also encode cyanophycinase and IaaA. Similarly, all the cphA1-harbouring Actinomycetota genomes also include cyanophycinase and iadA and/or iaaA. However, β-Proteobacteria genomes often encode CphA1 and IadA/IaaA, but not a recognizable cyanophycinase. Conversely, Firmicutes, Bacteroidota, α- and γ-Proteobacteria often encode CphA1 and cyanophycinase, but not IadA, nor IaaA.
Since there has been a strong research attention on cyanophycin metabolism within cyanobacteria, we also checked whether cphA1 co-occurs in cyanobacterial genomes with nifHDK, which encode the core components of nitrogenase40,41, and asr1734, a biomarker for heterocyst formation42. Around half of the CphA1-encoding, complete cyanobacterial genomes also contain all three of nifH, nifD and nifK (37/79 genomes), and 29 of them also contain asr1734, consistent with cyanophycin metabolism being useful for heterocyst-forming, nitrogen-fixing cyanobacteria8.
Identification of cyanophycin-metabolizing gene clusters
Next, we examined the tendency of cyanophycin-metabolizing genes to cluster together. We defined clustering as genes separated by not more than a 5 kilobase pair (kbp) intergenic region. Of the 658 genomes that have cphA1 and recognizable cyanophycinase, these genes are clustered in most (535; 81%; Tables 1, 3). However, in contrast to cyanophycinase genes, isoaspartyl dipeptidase genes generally do not cluster with cphA1, being proximal in only 88 (7.5%) of genomes that have both (Tables 1, 3).
Interestingly, clustering of cphA1 and isoaspartyl dipeptidase is more common in genomes that have genes encoding all three steps of cyanophycin metabolism (Tables 1, 3). There are 366 such genomes in the NCBI RefSeq database at time of analysis. In genomes that have cphA1, a cyanophycinase gene, and iaaA, 49 of 153 show clustering. In the case of iadA, 37 of 231 genomes with cphA1, a cyanophycinase gene and iadA show these three clustered.
Ben Hania et al. have described the utililty and occurance of a “cyanophycin utilization locus” which includes cyanophycinase genes, iadA and a transporter so a microbe can scavenge cyanophycin from the environment43. This observation also holds for iaaA: Searches of the NCBI RefSeq database returned 52 genomes that contain a cyanophycinase gene and iaaA or iadA but not cphA1, and 45 of them had cyanophycinase and isoaspartyl-dipeptidase genes clustered (Table 4).
The rate of each of the above clusterings is above random chance: As a control, we detected 955 genomes with cphA1 and dihydrofolate reductase (folA), a common housekeeping gene unrelated to cyanophycin metabolism. None of these genomes had the two genes clustered together (Table S3).
IadA and IaaA from cyanophycin clusters are not specific for β-Asp-Arg/Lys
Previous studies which characterized the activity of canonical isoaspartyl dipeptidases found that both IadA27,32 and IaaA20 accept a wide range of β-aspartyl dipeptides as substrates. Subsequent structures of the enzymes from E. coli32,44, which does not possess cyanophycin metabolizing genes, explained this lack of substrate specificity: while both enzymes make extensive interactions with the Asp portion of the substrate, the portion of the isoaspartyl dipeptidase surrounding the amino acid attached to the Asp side chain is large and able to accommodate the substrate rather than bind it specifically32,44.
We wondered whether the IaaA or IadA homologs present in cyanophycin metabolism clusters have evolved to specialize in cyanophycin degradation and display substrate preference for β-Asp-Arg (and β-Asp-Lys) over other β-aspartyl dipeptides. We therefore performed biochemical and structural characterization of a representative of IaaA and of IadA β-aspartyl dipeptidases whose genes are clustered with both cphA1 and cphB: IadA from the γ-proteobacteria Leucothrix mucor DSM2157 (LmIadA) and IaaA from the α-proteobacteria Roseivivax halodurans DSM15395 (RhIaaA).
LmIadA has 44% sequence identity to E. coli IadA (EcIadA32). Like EcIadA, the purified enzyme forms octamers in solution (Supplementary Fig. S2)32. We examined the activity of LmIadA towards several β-aspartyl dipeptides and found that it displayed no apparent preference towards β-Asp-Arg/Lys (Fig. 2a). To confirm the structural basis for this lack of specificity, we solved the structure of the wild type enzyme at 1.8 Å resolution and compared it to that of EcIadA32 (Table S1).
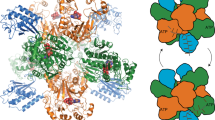
Structure and activity of LmIadA. (a) Asp release assay of LmIadA and different Asp-containing dipeptides. The enzyme is specific towards β-aspartyl dipeptides, but displays no specificity towards Arg or Lys as the β-linked amino acid. Individual measurements are shown as outlined white dots. Error bars represent the standard deviation of the mean of n = 4 replicates. (b) The homooctameric crystal structure of LmIadA. (c) Overlay of LmIadA (purple) and EcIadA32 (cyan, PDB code 1YBQ) monomers showing their high overall structural similarity. (d) Close-up view of the active sites of LmIadA and EcIadA in complex with the substrate β-Asp-His, showing they are similar in both sequence and structure. (e) Overlay of the regions around the active sites of LmIadA and EcIadA, showing both have large openings capable of accommodating a variety of β-aspartyl dipeptides as substrate.
The crystal structure of LmIadA shows a homooctameric architecture as the asymmetric unit (Fig. 2b). It displays high similarity to that of EcIadA32 (0.81 Å RMSD across 315 Cα pairs, PDB code 1YBQ; Fig. 2c), with the active site residues almost identical in both sequence and structure (Fig. 2d). Two Zn2+ ions are liganded by H64, H66, H198, H227 and E159, corresponding to EcIadA H68, H70, H201, H230 and carboxylated K162. Substrate binding residues in EcIadA such as E77, T106 and R23332, are also present at corresponding positions in LmIadA (E73, T102 and R230) and display similar conformations (Fig. 2d).
The published structure of EcIadA in complex with β-Asp-His32 shows that the His side chain of the substrate forms minimal interactions with the enzyme. It faces an opening in the active site which, as expected, can accommodate a variety of substrates. LmIadA displays a somewhat different architecture in this region (Fig. 2e). The loop formed by LmIadA T251-Y262 is longer and bulkier than the corresponding one of EcIadA (S255-V261), and as a result could restrict access to the active site. However, the partially flexible region between G288-G306 (EcIadA G288-G303) is oriented away from the binding pocket. This leads to a similarly sized opening in the active site region surrounding the non-Asp portion of the substrate and explains the lack of specificity (Fig. 2e).
We performed analogous analyses with the IaaA enzyme encoded in the cyanophycin gene cluster of Roseivivax halodurans. RhIaaA has 51% sequence identity with E. coli IaaA (EcIaaA44). Like EcIaaA and other Ntn-family enzymes, the pro-enzyme is expressed as a single chain that undergoes autocatalytic cleavage into two subunits, a and b, which constitute the mature a2b2 heterotetrameric enzyme (Supplementary Fig. S2). We assayed the activity of RhIaaA towards the same set of β-Asp dipeptides used to assess LmIadA and found that it could hydrolyze all of them with no apparent preference towards β-Asp-Arg/Lys (Fig. 3a). To confirm the structural basis for the lack of substrate specificity, we solved the structure of the wildtype enzyme at 2.7 Å resolution and compared it to that of EcIaaA (Supplementary Table S1).
Structure and activity of RhIaaA. (a) Asp release assay of RhIaaA with different Asp-containing dipeptides. The enzyme is specific towards β-aspartyl dipeptides, but displays no specificity towards Arg or Lys as the β-linked amino acid. Individual measurements are shown as outlined white dots. Error bars represent the standard deviation of the mean of n = 4 replicates. (b) The heterotetrameric crystal structure of RhIaaA. (c) Overlay of RhIaaA (purple) and EcIaaA44 (gray, PDB code 2ZAL) heterodimers showing their high overall structural similarity. (d) Close view of the active sites of RhIaaA and EcIaaA in complex with the product Asp, showing they are similar in both sequence and structure.
The crystal structure of RhIaaA shows the expected heterotetrameric architecture (Fig. 3b). The enzyme displays high structural similarity to EcIaaA (0.58 Å RMSD across 230 Cα pairs, PDB code 2ZAL44; Fig. 3c), with the active site residues being almost identical in both sequence and conformation. In EcIaaA, the Asp portion of the substrate is bound by T197, R207, D210, S211, T230 and G231, as well as the catalytic T17944. These residues are all present and in the same conformations in RhIaaA (T183, R193, D196, S197, T216 and G217, and the catalytic T165, Fig. 3d). As is the case with IadA, the substrate likely binds oriented in a way that positions the non-Asp portion of it facing a large opening in the active site (Fig. 3d). This presumably results in minimal interaction between IaaA and the substrate residue bound to Asp by the scissile isopeptide bond, which would enable the active site to accommodate a wide range of β-aspartyl dipeptides.
Discussion
Bacteria often use clustering to control expression of genes with related functions45. In the case of cyanophycin metabolism, clustering appears to be common for cphA1 and cyanophycinase9 (Table 1). Previous studies in cyanobacteria show that these two genes can also share some transcription regulation elements9. Clustering of genes for cyanophycinase and an isoaspartyl dipeptidase is very common in genomes that have those genes but not cphA1 (Table 2). These are often accompanied by amino acid transporters and probably represent cyanophycin-scavenging clusters, such as the ones described in the cyanobacteria-scavenger strain L21-Spi-D443 and in Flammeovirga pacifica strain WPAGA146.
The clustering rate of isoaspartyl dipeptidases with cphA1 and cyanophycinase in genomes that have all three is well above random distribution, but not as high as that of the cphA1-cyanophycinase pair. There are several possible explanations why clustering is not strict. First, it is possible for these genes to be under control of the same transcription regulators even if they are not clustered. Second, since isoaspartyl dipeptidases are required outside of a cyanophycin context, there may be evolutionary pressure to keep those genes separate for regulatory purposes. Third, in some cases it is beneficial to have cyanophycin-metabolizing genes regulated independently of one another. An example for this can be seen in the heterocyst-forming cyanobacterium Anabaena sp. PCC7120. Heterocysts of this bacterium express cyanophycinase to degrade cyanophycin into dipeptides, which are shuttled to vegetative cells. These, in turn, express high levels of IaaA to convert the dipeptides into free amino acids8.
The co-occurrence rates of genes involved in cyanophycin metabolism show distinct patterns in different clades. For example, detection of a recognizable cyanophycinase in only 3% of cphA1-containing β-Proteobacterial genomes is unanticipated. Cyanophycin is only known to serve as a storage material, so the bacteria that store it must also possess the wherewithal to degrade it. It is possible that bacteria which possess cphA1 but not cphB/E/I possess other, unknown cyanophycinase isozymes. The lack of an identifiable isoaspartyl dipeptidase gene in many Firmicutes, Bacteroidota, α- and γ-Proteobacteria in cphA1-containing genomes suggests that not all genes encoding enzymes with this dipeptidase activity were detected in our searches. Similarly, Füser et al. performed an analysis of 48 cphA1 or cyanophycinase-containing genomes in 20077 and found that only 26 also had iaaA or iadA. Isoaspartyl dipeptidase activity in these bacteria could be provided by distant homologues of iaaA or iadA or by unrelated isozymes. The existence of cryptic isoaspartyl dipeptidase enzymes has been proposed before, for example in Saccharomyces cerevisiae47. Manual examination of genomes from the NCBI RefSeq database that have a CphA1-cyanophycinase cluster shows some of them to include adjacent genes which could potentially have isoaspartyl dipeptidase activity, such as those annotated as “S9 family peptidase” (in genome NZ_CP029187.1), annotated as “M14 family metallopeptidase” or “succinylglutamate desuccinylase/aspartoacylase family protein” (in genome NZ_VYQF01000002.1) and a gene weakly homologous (25–30% identity) to cocaine esterase48 (in genome NZ_SJEY01000003). Indeed, during the publication process of these results, a study which we initiated after this one shows that the enzymes annotated as “M14 family metallopeptidase” or “succinylglutamate desuccinylase/aspartoacylase family protein” are isoaspartyl dipeptidases with specificity for β-Asp-Arg and β-Asp-Lys49. This novel “cyanophycin dipeptide hydrolase” family explains the missing isoaspartyl dipeptidase in α- and γ-Proteobacteria49, but the isoaspartyl dipeptidase(s) in Firmicutes and Bacteroidota remain undiscovered.
Both of the isoaspartyl dipeptidases from cyanophycin gene clusters that we cloned, expressed, purified and assayed display no substrate specificity towards β-Asp-Arg/Lys and accept a range of isoaspartyl dipeptides. The crystal structures of both enzymes were consistent with this promiscuity and show that the structural basis for this lack of specificity is shared with other IaaA and IadA enzymes. Although clustered genes are not necessarily co-regulated, and distal genes can be co-regulated, these results suggest that even when their genes are clustered with cyanophycin-related genes, IaaA and IadA function in both cyanophycin metabolism and the protein-degradation pathway. This assertion is in line with the widely held belief that general isoaspartyl dipeptidases are usually49 responsible for the last step of cyanophycin degradation7,20.
Methods
Bioinformatics
For the identification of gene clusters, we created a local database with all complete bacterial genomes in the NCBI (USA) Refseq39 database (May 2022). We used cblaster50 to search this database using several queries for CphA1 (Synechocystis sp. PCC 6714 WP_028947105.1, Acinetobacter baylyi ADP1 WP_004925893.1, Desulfitobacterium hafniense DCB-2 WP_015942562.1), cyanophycinase (Synechococcus elongatus WP_011058003.1, Acinetobacter baylyi ADP1 WP_004925892.1, Pseudomonas anguilliseptica B1 Q8KQN8.1), IadA (Pedobacter mendelii CCM 8939 WP_188415469.1, Caloramator sp. E03 WP_138978951.1, Roseivivax halodurans JCM 10272 WP_037265155.1), IaaA (Candidatus Bipolaricaulota MBS3792760.1, Ezakiella coagulans DSM 20705 WP_034545427.1, Leucothrix mucor DSM 2157 WP_022952024.1), NifH (Nostoc sp. PCC7120 WP_010995626.1), NifD (Nostoc sp. PCC 7120 WP_044520961.1), NifK (Nostoc sp. PCC7120 WP_010995612.1) and Asr1734 (Trichormus variabilis ATCC 29413 WP_010995902.1). For the identification of putative isoaspartyl dipeptidases in cphA1-cyanophycinase clusters, MultiGeneBlast51 was used to search cphA1-containing genomes for cphA1-cyanophycinase clusters, and the results were analyzed manually for putative isoaspartyl dipeptidases. Sorting of hits by bacterial clade was performed with PhyloT and iTOL52 using the Genome Taxonomy Database53 phylogenetic tree.
Cloning, protein expression and purification
The genes encoding LmIadA (WP_022952024.1) and RhIaaA (WP_037265155.1) were amplified from genomic DNA (DSMZ, Leibniz Institute, Germany). Both genes were cloned into a plasmid derived from pJ411 with a C-terminal tobacco etch virus (TEV) protease cleavage site and an 8xHis affinity tag. Gene subcloning and mutagenesis were performed by transforming PCR fragments with overlapping ends into chemically competent DH5-α E. coli cells. Proteins were expressed in E. coli BL21(DE3) cells grown in TB media supplemented with 150 µg/ml kanamycin. Cultures were grown at 37 °C until they reached an OD600 of ~ 1. The growth temperature was then lowered to 18 °C and protein expression was induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for ~ 20 h. All subsequent protein purification steps were carried out at 4 °C. Following harvest by centrifugation, the cells were resuspended in buffer A (250 mM NaCl, 50 mM Tris pH 8.0, 10 mM imidazole, 2 mM β-mercaptoethanol) supplemented with a few crystals of lysozyme and DNase I, and lysed by sonication. The lysate was clarified by centrifugation at 40,000 g for 30 min and then applied onto a HisTrap HP column (Cytiva, USA). The column was washed extensively with buffer B (buffer A with 30 mM imidazole) and the protein was eluted with buffer C (buffer A with 250 mM imidazole). For structural studies, the protein was incubated with TEV protease for removal of the 8xHis tag while being dialyzed overnight against buffer D (250 mM NaCl, 20 mM Tris pH 8.0, 5 mM β-mercaptoethanol) prior to application to a HisTrap column and collection of the flow through. All protein preparations were then concentrated and applied to a Superdex200 16/60 column (Cytiva, USA) equilibrated in buffer E (100 mM NaCl, 20 mM Tris pH 8.0, 1 mM dithiothreitol). Fractions with the highest protein purity were concentrated, supplemented with glycerol to a final volume of 15% and flash frozen in liquid nitrogen for storage.
Protein crystallization, data collection, structure solution and refinement
For crystallization trials, all proteins were buffer exchanged into buffer E and subjected to small-scale wide screen crystallization trials in 96-well plates using the sitting drop method. Optimization of crystallization conditions was performed using the sitting drop method by mixing 2 µl of protein with 2 µl of crystallization buffer and allowing this to equilibrate against 500 µl of crystallization buffer. The crystallization buffer for LmIadA (20 mg/ml) contained 0.56 M NaH2PO4 and 1.04 M K2HPO4. Crystals were grown at 22 °C and cryo-protected by briefly dipping them in crystallization solution supplemented with 20% glycerol before freezing in liquid nitrogen. Data were collected at the Advanced Light Source (ALS) beamline 5.0.1. The structure was solved by molecular replacement using E. coli IadA (PDB code 1YBQ) as a search model. The crystallization buffer for RhIaaA (10 mg/ml) contained 0.1 M bis–tris propane pH 8.5, 0.2 M disodium malonate and 25% PEG3350. Crystals were grown at 4 °C and cryo-protected by dipping them in crystallization solution supplemented with 10% PEG100 for 1 min before freezing in liquid nitrogen. Data were collected at the Canadian Light Source (CLS) beamline CMCF-BM. The structure was solved by molecular replacement using E. coli IaaA (PDB code 2ZAL) as a search model. All datasets were processed in DIALS54 and merged in AIMLESS55 implemented in CCP4i2 suite56. The structures were refined in REFMAC557, Rosetta58, Phenix59 and Coot60. Figures were prepared in PyMOL (Schrödinger, USA).
Enzyme activity assays
Enzyme-catalyzed β-Asp-X dipeptide hydrolysis was measured with an Asp release assay32. The 100 µl reactions contained 100 mM HEPES pH 8.2, 20 mM KCl, 5 mM α-ketoglutarate, 1 mM NADH, 2.4 U aspartate aminotransferase, 0.3 U malate dehydrogenase, 1 mM dipeptide substrate and 500 nM purified enzyme. Data were collected by following 340 nm transmittance in 96-well plates using a SpectraMax Paradigm (Molecular Devices, USA) and analyzed using Prism (GraphPad, USA). β-Asp-Arg dipeptides were purified as previously described18. β-Asp-Ala and α-Asp-Arg were purchased from Bachem (Switzerland). β-Asp-Lys and β-Asp-Leu were purchased from Toronto Research Chemicals (Canada). β-Asp-Asp was purchased from Advanced ChemBlocks (USA).
Data availability
Diffraction data and structures determined in this study have been deposited to the Protein Data Bank: LmIadA (PDB 8DQN; https://www.rcsb.org/structure/8DQN), RhIaaA (PDB 8DQM https://www.rcsb.org/structure/8DQM). All other relevant data are within the manuscript and its Supplementary Information files.
References
Borzi, A. Le communicazioni intracellulari delle nostochinee. Malpighia - Rassegna mensuale di botanica 1 (1886–1887).
Simon, R. D. Cyanophycin granules from the blue-green alga anabaena cylindrica: A reserve material consisting of copolymers of aspartic acid and arginine. Proc. Natl. Acad. Sci. U. S. A. 68, 265–267 (1971).
Liotenberg, S., Campbell, D., Rippka, R., Houmard, J. & de Marsac, N. T. Effect of the nitrogen source on phycobiliprotein synthesis and cell reserves in a chromatically adapting filamentous cyanobacterium. Microbiology 142, 611–622. https://doi.org/10.1099/13500872-142-3-611 (1996).
Liang, B. et al. Cyanophycin mediates the accumulation and storage of fixed carbon in non-heterocystous filamentous cyanobacteria from coniform mats. PLoS One 9, e88142. https://doi.org/10.1371/journal.pone.0088142 (2014).
Wingard, L. L. et al. Cyanophycin production in a phycoerythrin-containing marine synechococcus strain of unusual phylogenetic affinity. Appl. Environ. Microbiol. 68, 1772–1777. https://doi.org/10.1128/aem.68.4.1772-1777.2002 (2002).
Sharon, I. et al. Structures and function of the amino acid polymerase cyanophycin synthetase. Nat. Chem. Biol. 17, 1101–1110. https://doi.org/10.1038/s41589-021-00854-y (2021).
Fuser, G. & Steinbuchel, A. Analysis of genome sequences for genes of cyanophycin metabolism: Identifying putative cyanophycin metabolizing prokaryotes. Macromol. Biosci. 7, 278–296. https://doi.org/10.1002/mabi.200600207 (2007).
Burnat, M., Herrero, A. & Flores, E. Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. U. S. A. 111, 3823–3828. https://doi.org/10.1073/pnas.1318564111 (2014).
Picossi, S., Valladares, A., Flores, E. & Herrero, A. Nitrogen-regulated genes for the metabolism of cyanophycin, a bacterial nitrogen reserve polymer: Expression and mutational analysis of two cyanophycin synthetase and cyanophycinase gene clusters in heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. J. Biol. Chem. 279, 11582–11592 (2004).
Simon, R. D. The biosynthesis of multi-L-arginyl-poly(L-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 422, 407–418 (1976).
Allen, M. M. & Weathers, P. J. Structure and composition of cyanophycin granules in the cyanobacterium Aphanocapsa 6308. J. Bacteriol. 141, 959–962 (1980).
Mackerras, A. H., de Chazal, N. M. & Smith, G. D. Transient accumulations of cyanophycin in Anabaena cylindrica and Synechocystis 6308. Microbiology 136, 2057–2065. https://doi.org/10.1099/00221287-136-10-2057 (1990).
Li, H., Sherman, D. M., Bao, S. & Sherman, L. A. Pattern of cyanophycin accumulation in nitrogen-fixing and non-nitrogen-fixing cyanobacteria. Arch. Microbiol. 176, 9–18. https://doi.org/10.1007/s002030100281 (2001).
Ziegler, K. et al. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-L-arginyl-poly-L-aspartate (cyanophycin). Eur. J. Biochem. 254, 154–159 (1998).
Klemke, F. et al. CphA2 is a novel type of cyanophycin synthetase in N2-fixing cyanobacteria. Microbiology 162, 526–536 (2016).
Sharon, I. et al. A cryptic third active site in cyanophycin synthetase creates primers for polymerization. Nat. Commun. 13, 3923. https://doi.org/10.1038/s41467-022-31542-7 (2022).
Frommeyer, M. & Steinbuchel, A. Increased lysine content is the main characteristic of the soluble form of the polyamide cyanophycin synthesized by recombinant Escherichia coli. Appl. Environ. Microbiol. 79, 4474–4483. https://doi.org/10.1128/AEM.00986-13 (2013).
Sharon, I., Grogg, M., Hilvert, D. & Schmeing, T. M. Structure and function of the β-Asp-Arg polymerase cyanophycin synthetase 2. ACS Chem. Biol. 17, 670–679 (2022).
Richter, R., Hejazi, M., Kraft, R., Ziegler, K. & Lockau, W. Cyanophycinase, a peptidase degrading the cyanobacterial reserve material multi-L-arginyl-poly-L-aspartic acid (cyanophycin): Molecular cloning of the gene of Synechocystis sp. PCC 6803, expression in Escherichia coli, and biochemical characterization of the purified enzyme. Eur. J. Biochem. 263, 163–169 (1999).
Hejazi, M. et al. Isoaspartyl dipeptidase activity of plant-type asparaginases. Biochem. J. 364, 129–136 (2002).
Watzer, B. & Forchhammer, K. Cyanophycin synthesis optimizes nitrogen utilization in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.01298-18 (2018).
Sallam, A. & Steinbuchel, A. Anaerobic and aerobic degradation of cyanophycin by the denitrifying bacterium Pseudomonas alcaligenes strain DIP1 and role of three other coisolates in a mixed bacterial consortium. Appl. Environ. Microbiol. 74, 3434–3443. https://doi.org/10.1128/AEM.02575-07 (2008).
Obst, M., Oppermann-Sanio, F. B., Luftmann, H. & Steinbuchel, A. Isolation of cyanophycin-degrading bacteria, cloning and characterization of an extracellular cyanophycinase gene (cphE) from Pseudomonas anguilliseptica strain BI. The cphE gene from P. anguilliseptica BI encodes a cyanophycin hydrolyzing enzyme. J. Biol. Chem. 277, 25096–25105. https://doi.org/10.1074/jbc.M112267200 (2002).
Sallam, A. & Steinbuchel, A. Cyanophycin-degrading bacteria in digestive tracts of mammals, birds and fish and consequences for possible applications of cyanophycin and its dipeptides in nutrition and therapy. J. Appl. Microbiol. 107, 474–484. https://doi.org/10.1111/j.1365-2672.2009.04221.x (2009).
Obst, M., Sallam, A., Luftmann, H. & Steinbuchel, A. Isolation and characterization of gram-positive cyanophycin-degrading bacteria-kinetic studies on cyanophycin depolymerase activity in aerobic bacteria. Biomacromolecules 5, 153–161. https://doi.org/10.1021/bm034281p (2004).
Obst, M., Krug, A., Luftmann, H. & Steinbuchel, A. Degradation of cyanophycin by Sedimentibacter hongkongensis strain KI and Citrobacter amalonaticus strain G isolated from an anaerobic bacterial consortium. Appl. Environ. Microbiol. 71, 3642–3652. https://doi.org/10.1128/AEM.71.7.3642-3652.2005 (2005).
Gary, J. D. & Clarke, S. Purification and characterization of an isoaspartyl dipeptidase from Escherichia coli. J. Biol. Chem. 270, 4076–4087. https://doi.org/10.1074/jbc.270.8.4076 (1995).
Radkiewicz, J. L., Zipse, H., Clarke, S. & Houk, K. N. Accelerated racemization of aspartic acid and asparagine residues via succinimide intermediates: An ab initio theoretical exploration of mechanism. J. Am. Chem. Soc. 118, 9148–9155. https://doi.org/10.1021/ja953505b (1996).
Kim, E., Lowenson, J. D., MacLaren, D. C., Clarke, S. & Young, S. G. Deficiency of a protein-repair enzyme results in the accumulation of altered proteins, retardation of growth, and fatal seizures in mice. Proc. Natl. Acad. Sci. U. S. A. 94, 6132–6137. https://doi.org/10.1073/pnas.94.12.6132 (1997).
Lowenson, J. D., Kim, E., Young, S. G. & Clarke, S. Limited accumulation of damaged proteins in l-isoaspartyl (D-aspartyl) O-methyltransferase-deficient mice. J. Biol. Chem. 276, 20695–20702. https://doi.org/10.1074/jbc.M100987200 (2001).
Aswad, D. W., Paranandi, M. V. & Schurter, B. T. Isoaspartate in peptides and proteins: Formation, significance, and analysis. J. Pharm. Biomed. Anal. 21, 1129–1136. https://doi.org/10.1016/s0731-7085(99)00230-7 (2000).
Marti-Arbona, R. et al. Mechanism of the reaction catalyzed by isoaspartyl dipeptidase from Escherichia coli. Biochemistry 44, 7115–7124. https://doi.org/10.1021/bi050008r (2005).
Borek, D. et al. Expression, purification and catalytic activity of Lupinus luteus asparagine beta-amidohydrolase and its Escherichia coli homolog. Eur. J. Biochem. 271, 3215–3226. https://doi.org/10.1111/j.1432-1033.2004.04254.x (2004).
Prahl, A., Pazgier, M., Hejazi, M., Lockau, W. & Lubkowski, J. Structure of the isoaspartyl peptidase with L-asparaginase activity from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 60, 1173–1176. https://doi.org/10.1107/S0907444904003403 (2004).
Flores, E., Arévalo, S. & Burnat, M. Cyanophycin and arginine metabolism in cyanobacteria. Algal Res. 42, 101577. https://doi.org/10.1016/j.algal.2019.101577 (2019).
Haley, E. E. Purification and properties of a β-aspartyl peptidase from Escherichia coli. J. Biol. Chem. 243, 5748–5752. https://doi.org/10.1016/S0021-9258(18)91928-9 (1968).
Law, A. M., Lai, S. W., Tavares, J. & Kimber, M. S. The structural basis of beta-peptide-specific cleavage by the serine protease cyanophycinase. J. Mol. Biol. 392, 393–404. https://doi.org/10.1016/j.jmb.2009.07.001 (2009).
Noronkoski, T., Stoineva, I. B., Ivanov, I. P., Petkov, D. D. & Mononen, I. Glycosylasparaginase-catalyzed synthesis and hydrolysis of beta-aspartyl peptides. J. Biol. Chem. 273, 26295–26297. https://doi.org/10.1074/jbc.273.41.26295 (1998).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. https://doi.org/10.1093/nar/gkv1189 (2016).
Chen, M. Y. et al. Phylogenomics uncovers evolutionary trajectory of nitrogen fixation in cyanobacteria. Mol. Biol. Evol. https://doi.org/10.1093/molbev/msac171 (2022).
Raymond, J., Siefert, J. L., Staples, C. R. & Blankenship, R. E. The natural history of nitrogen fixation. Mol. Biol. Evol. 21, 541–554. https://doi.org/10.1093/molbev/msh047 (2004).
Kumar, K., Mella-Herrera, R. A. & Golden, J. W. Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2, a000315. https://doi.org/10.1101/cshperspect.a000315 (2010).
Ben Hania, W. et al. Characterization of the first cultured representative of a Bacteroidetes clade specialized on the scavenging of cyanobacteria. Environ. Microbiol. 19, 1134–1148. https://doi.org/10.1111/1462-2920.13639 (2017).
Michalska, K., Brzezinski, K. & Jaskolski, M. Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate. J. Biol. Chem. 280, 28484–28491. https://doi.org/10.1074/jbc.M504501200 (2005).
Osbourn, A. E. & Field, B. Operons. Cell. Mol. Life Sci. 66, 3755–3775. https://doi.org/10.1007/s00018-009-0114-3 (2009).
Chan, Z. et al. Draft genome sequence of an agar-degrading marine bacterium Flammeovirga pacifica WPAGA1. Mar. Genom. 20, 23–24. https://doi.org/10.1016/j.margen.2014.12.001 (2015).
Patananan, A. N., Capri, J., Whitelegge, J. P. & Clarke, S. G. Non-repair pathways for minimizing protein isoaspartyl damage in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 289, 16936–16953. https://doi.org/10.1074/jbc.M114.564385 (2014).
Bresler, M. M., Rosser, S. J., Basran, A. & Bruce, N. C. Gene cloning and nucleotide sequencing and properties of a cocaine esterase from Rhodococcus sp. strain MB1. Appl. Environ. Microbiol. 66, 904–908. https://doi.org/10.1128/AEM.66.3.904-908.2000 (2000).
Sharon, I., McKay, G. A., Nguyen, D. & Schmeing, T. M. Discovery of cyanophycin dipeptide hydrolase enzymes suggests widespread utility of the natural biopolymer cyanophycin. Proc. Natl. Acad. Sci. U. S. A. 120, e2216547120. https://doi.org/10.1073/pnas.2216547120 (2023).
Gilchrist, C. L. M. et al. cblaster: A remote search tool for rapid identification and visualization of homologous gene clusters. Bioinform. Adv. 1, vbab016. https://doi.org/10.1093/bioadv/vbab016 (2021).
Medema, M. H., Takano, E. & Breitling, R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30, 1218–1223. https://doi.org/10.1093/molbev/mst025 (2013).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259. https://doi.org/10.1093/nar/gkz239 (2019).
Parks, D. H. et al. GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794. https://doi.org/10.1093/nar/gkab776 (2021).
Beilsten-Edmands, J. et al. Scaling diffraction data in the DIALS software package: Algorithms and new approaches for multi-crystal scaling. Acta Crystallogr. D Struct. Biol. 76, 385–399. https://doi.org/10.1107/S2059798320003198 (2020).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution?. Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214. https://doi.org/10.1107/S0907444913000061 (2013).
Potterton, L. et al. CCP4i2: The new graphical user interface to the CCP4 program suite. Acta Crystallogr. D Struct. Biol. 74, 68–84. https://doi.org/10.1107/S2059798317016035 (2018).
Kovalevskiy, O., Nicholls, R. A. & Murshudov, G. N. Automated refinement of macromolecular structures at low resolution using prior information. Acta Crystallogr. D Struct. Biol. 72, 1149–1161. https://doi.org/10.1107/S2059798316014534 (2016).
Song, Y. et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742. https://doi.org/10.1016/j.str.2013.08.005 (2013).
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106. https://doi.org/10.1016/j.ymeth.2011.07.005 (2011).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. https://doi.org/10.1107/S0907444904019158 (2004).
Acknowledgements
We thank all the members of the Schmeing lab for important advice and ongoing discussions on this project, Nancy Rogerson for proofreading and synchrotron staff J. Gorin (Canadian Light Source) and M. Allaire (Advanced Light Source) for facilitating remote collection of diffraction datasets. Part of the research described in this paper was performed using beamline CMCF-BM at the Canadian Light Source, a national research facility of the University of Saskatchewan, which is supported by the Canada Foundation for Innovation (CFI), the Natural Sciences and Engineering Research Council (NSERC), the National Research Council (NRC), the Canadian Institutes of Health Research (CIHR), the Government of Saskatchewan, and the University of Saskatchewan. Beamline 5.0.1 of the Advanced Light Source, a U.S. DOE Office of Science User Facility under Contract No. DE-AC02-05CH11231, is supported in part by the ALS-ENABLE program funded by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01. This work was funded by CIHR Project Grant 178084 and a Canada Research Chair to TMS.
Author information
Authors and Affiliations
Contributions
I.S. and T.M.S. designed the study and performed the bioinformatic analysis. I.S. performed all biochemical and structural experiments and data processing. I.S. and T.MS. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharon, I., Schmeing, T.M. Bioinformatics of cyanophycin metabolism genes and characterization of promiscuous isoaspartyl dipeptidases that catalyze the final step of cyanophycin degradation. Sci Rep 13, 8314 (2023). https://doi.org/10.1038/s41598-023-34587-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34587-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.