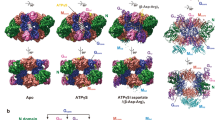
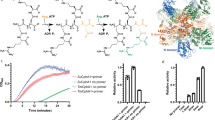
Abstract
Cyanophycin is a natural biopolymer produced by a wide range of bacteria, consisting of a chain of poly-l-Asp residues with l-Arg residues attached to the β-carboxylate sidechains by isopeptide bonds. Cyanophycin is synthesized from ATP, aspartic acid and arginine by a homooligomeric enzyme called cyanophycin synthetase (CphA1). CphA1 has domains that are homologous to glutathione synthetases and muramyl ligases, but no other structural information has been available. Here, we present cryo-electron microscopy and X-ray crystallography structures of cyanophycin synthetases from three different bacteria, including cocomplex structures of CphA1 with ATP and cyanophycin polymer analogs at 2.6 Å resolution. These structures reveal two distinct tetrameric architectures, show the configuration of active sites and polymer-binding regions, indicate dynamic conformational changes and afford insight into catalytic mechanism. Accompanying biochemical interrogation of substrate binding sites, catalytic centers and oligomerization interfaces combine with the structures to provide a holistic understanding of cyanophycin biosynthesis.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The cryo-EM maps created in this study have been deposited to the EMDB: SuCphA1 bound with ATP (EMD-23311), SuCphA1 bound with ADPCP and (β-Asp-Arg)8-NH2 (EMD-23325), SuCphA1 bound with ATP and (β-Asp-Arg)8-Asn (EMD-23328), SuCphA1 with ATP and (β-Asp-Arg)16 (EMD-23326) and AbCphA1 with ATP (EMD-23327). The protein structures solved in this study have been deposited to the PDB: SuCphA1 with ATP (7LG5), SuCphA1 with ADPCP and (β-Asp-Arg)8-NH2 (7LGJ), SuCphA1 with ATP and (β-Asp-Arg)8-Asn (7LGQ), SuCphA1 with ATP, AbCphA1 with ATP (7LGM) and TmCphA1 (7LGN). Source data are provided with this paper.
References
Borzi, A. Le communicazioni intracellulari delle nostochinee (Messina, 1886).
Simon, R. D. The biosynthesis of multi-l-arginyl-poly(l-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 422, 407–418 (1976).
Simon, R. D. Cyanophycin granules from the blue-green alga Anabaena cylindrica: a reserve material consisting of copolymers of aspartic acid and arginine. Proc. Natl Acad. Sci. USA 68, 265–267 (1971).
Liotenberg, S., Campbell, D., Rippka, R., Houmard, J. & de Marsac, N. T. Effect of the nitrogen source on phycobiliprotein synthesis and cell reserves in a chromatically adapting filamentous cyanobacterium. Microbiology 142, 611–622 (1996).
Mariotti, F., Tome, D. & Mirand, P. P. Converting nitrogen into protein–beyond 6.25 and Jones’ factors. Crit. Rev. Food Sci. Nutr. 48, 177–184 (2008).
Li, H., Sherman, D. M., Bao, S. & Sherman, L. A. Pattern of cyanophycin accumulation in nitrogen-fixing and non-nitrogen-fixing cyanobacteria. Arch. Microbiol. 176, 9–18 (2001).
Fay, P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev. 56, 340–373 (1992).
Kumar, K., Mella-Herrera, R. A. & Golden, J. W. Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2, a000315 (2010).
Sukenik, A. et al. Carbon assimilation and accumulation of cyanophycin during the development of dormant cells (akinetes) in the cyanobacterium Aphanizomenon ovalisporum. Front. Microbiol. 6, 1067 (2015).
Burnat, M., Herrero, A. & Flores, E. Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium. Proc. Natl Acad. Sci. USA 111, 3823–3828 (2014).
Liang, B. et al. Cyanophycin mediates the accumulation and storage of fixed carbon in non-heterocystous filamentous cyanobacteria from coniform mats. PLoS ONE 9, e88142 (2014).
Wingard, L. L. et al. Cyanophycin production in a phycoerythrin-containing marine synechococcus strain of unusual phylogenetic affinity. Appl. Environ. Microbiol. 68, 1772–1777 (2002).
Obst, M., Oppermann-Sanio, F. B., Luftmann, H. & Steinbuchel, A. Isolation of cyanophycin-degrading bacteria, cloning and characterization of an extracellular cyanophycinase gene (cphE) from Pseudomonas anguilliseptica strain BI. The cphE gene from P. anguilliseptica BI encodes a cyanophycinhydrolyzing enzyme. J. Biol. Chem. 277, 25096–25105 (2002).
Stevens, S. E. & Paone, D. A. Accumulation of cyanophycin granules as a result of phosphate limitation in Agmenellum quadruplicatum. Plant Physiol. 67, 716–719 (1981).
Liu, H., Ray, W. K., Helm, R. F., Popham, D. L. & Melville, S. B. Analysis of the spore membrane proteome in Clostridium perfringens implicates cyanophycin in spore assembly. J. Bacteriol. 198, 1773–1782 (2016).
Gorelova, O. A. & Kleimenov, S. The accumulation and degradation dynamics of cyanophycin in cyanobacteria grown in symbiotic associations with plant tissues and cells. Mikrobiologiia 72, 361–369 (2003).
Korzhenevskaya, T. G., Gorelova, O. A., Baulina, O. I. & Gusev, M. V. Accumulation of reserve polymers by Nostoc muscorum CALU 304 cells grown in mixed culture with plant tissue. Mikrobiologiia 68, 191–197 (1999).
Aboulmagd, E., Voss, I., Oppermann-Sanio, F. B. & Steinbuchel, A. Heterologous expression of cyanophycin synthetase and cyanophycin synthesis in the industrial relevant bacteria Corynebacterium glutamicum and Ralstonia eutropha and in Pseudomonas putida. Biomacromolecules 2, 1338–1342 (2001).
Nausch, H. et al. Tobacco as platform for a commercial production of cyanophycin. N. Biotechnol. 33, 842–851 (2016).
Huhns, M. et al. Plastid targeting strategies for cyanophycin synthetase to achieve high-level polymer accumulation in Nicotiana tabacum. Plant Biotechnol. J. 6, 321–336 (2008).
Gross, R. A. & Kalra, B. Biodegradable polymers for the environment. Science 297, 803–807 (2002).
Tseng, W. C., Fang, T. Y., Lin, Y. C., Huang, S. J. & Huang, Y. H. Reversible self-assembly nanovesicle of UCST response prepared with multi-l-arginyl-poly-l-aspartate conjugated with polyethylene glycol. Biomacromolecules 19, 4585–4592 (2018).
Ziegler, K. et al. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartate (cyanophycin). Eur. J. Biochem. 254, 154–159 (1998).
Hai, T., Oppermann-Sanio, F. B. & Steinbuchel, A. Molecular characterization of a thermostable cyanophycin synthetase from the thermophilic cyanobacterium Synechococcus sp. strain MA19 and in vitro synthesis of cyanophycin and related polyamides. Appl. Environ. Microbiol. 68, 93–101 (2002).
Arai, T. & Kino, K. A cyanophycin synthetase from Thermosynechococcus elongatus BP-1 catalyzes primer-independent cyanophycin synthesis. Appl. Microbiol. Biotechnol. 81, 69–78 (2008).
Hara, T., Kato, H., Katsube, Y. & Oda, J. A pseudo-Michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution. Biochemistry 35, 11967–11974 (1996).
Berg, H. et al. Biosynthesis of the cyanobacterial reserve polymer multi-l-arginyl-poly-l-aspartic acid (cyanophycin): mechanism of the cyanophycin synthetase reaction studied with synthetic primers. Eur. J. Biochem. 267, 5561–5570 (2000).
van Heijenoort, J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18, 503–519 (2001).
Hai, T., Frey, K. M. & Steinbuchel, A. Engineered cyanophycin synthetase (CphA) from Nostoc ellipsosporum confers enhanced CphA activity and cyanophycin accumulation to Escherichia coli. Appl. Environ. Microbiol. 72, 7652–7660 (2006).
Neubauer, K. et al. Isolation of cyanophycin from tobacco and potato plants with constitutive plastidic cphATe gene expression. J. Biotechnol. 158, 50–58 (2012).
Fuser, G. & Steinbuchel, A. Analysis of genome sequences for genes of cyanophycin metabolism: identifying putative cyanophycin metabolizing prokaryotes. Macromol. Biosci. 7, 278–296 (2007).
Laranjo, M., Alexandre, A. & Oliveira, S. Legume growth-promoting rhizobia: an overview on the Mesorhizobium genus. Microbiol. Res. 169, 2–17 (2014).
Zehr, J. P. & Ward, B. B. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68, 1015–1024 (2002).
Mota, C., Head, M. A., Ridenoure, J. A., Cheng, J. J. & de Los Reyes, F. L. 3rd Effects of aeration cycles on nitrifying bacterial populations and nitrogen removal in intermittently aerated reactors. Appl. Environ. Microbiol. 71, 8565–8572 (2005).
Watzer, B. & Forchhammer, K. Cyanophycin synthesis optimizes nitrogen utilization in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Appl. Environ. Microbiol. 84, e01298–18 (2018).
Bolotin, E. & Hershberg, R. Bacterial intra-species gene loss occurs in a largely clocklike manner mostly within a pool of less conserved and constrained genes. Sci. Rep. 6, 35168 (2016).
Krehenbrink, M. & Steinbuchel, A. Partial purification and characterization of a non-cyanobacterial cyanophycin synthetase from Acinetobacter calcoaceticus strain ADP1 with regard to substrate specificity, substrate affinity and binding to cyanophycin. Microbiology 150, 2599–2608 (2004).
Diaz-Saez, L., Torrie, L. S., McElroy, S. P., Gray, D. & Hunter, W. N. Burkholderia pseudomallei d-alanine-d-alanine ligase; detailed characterisation and assessment of a potential antibiotic drug target. FEBS J. 286, 4509–4524 (2019).
Hibi, T. et al. Structure of the multifunctional loops in the nonclassical ATP-binding fold of glutathione synthetase. Nat. Struct. Biol. 3, 16–18 (1996).
Stout, J., De Vos, D., Vergauwen, B. & Savvides, S. N. Glutathione biosynthesis in bacteria by bifunctional GshF is driven by a modular structure featuring a novel hybrid ATP-grasp fold. J. Mol. Biol. 416, 486–494 (2012).
Yamaguchi, H. et al. Three-dimensional structure of the glutathione synthetase from Escherichia coli B at 2.0 A resolution. J. Mol. Biol. 229, 1083–1100 (1993).
Li, H., Fast, W. & Benkovic, S. J. Structural and functional modularity of proteins in the de novo purine biosynthetic pathway. Protein Sci. 18, 881–892 (2009).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Galant, A., Arkus, K. A., Zubieta, C., Cahoon, R. E. & Jez, J. M. Structural basis for evolution of product diversity in soybean glutathione biosynthesis. Plant Cell 21, 3450–3458 (2009).
Gordon, E. et al. Crystal structure of UDP-N-acetylmuramoyl-l-alanyl-d-glutamate: meso-diaminopimelate ligase from Escherichia coli. J. Biol. Chem. 276, 10999–11006 (2001).
Smith, C. A. Structure, function and dynamics in the mur family of bacterial cell wall ligases. J. Mol. Biol. 362, 640–655 (2006).
Basavannacharya, C., Robertson, G., Munshi, T., Keep, N. H. & Bhakta, S. ATP-dependent MurE ligase in Mycobacterium tuberculosis: biochemical and structural characterisation. Tuberculosis (Edinb.) 90, 16–24 (2010).
Hai, T., Lee, J.-S., Kim, T.-J. & Suh, J.-W. The role of the C-terminal region of cyanophycin synthetase from Nostoc ellipsosporum NE1 in its enzymatic activity and thermostability: a key function of Glu(856). Biochim. Biophys. Acta 1794, 42–49 (2009).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Beilsten-Edmands, J. et al. Scaling diffraction data in the DIALS software package: algorithms and new approaches for multi-crystal scaling. Acta Crystallogr. D. Struct. Biol. 76, 385–399 (2020).
Foadi, J. et al. Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 69, 1617–1632 (2013).
Song, Y. et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Skubak, P., Murshudov, G. N. & Pannu, N. S. Direct incorporation of experimental phase information in model refinement. Acta Crystallogr. D. Biol. Crystallogr. 60, 2196–2201 (2004).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zhang, K. GCTF: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Grogg, M. et al. Cell penetration, herbicidal activity, and in-vivo-toxicity of oligo-arginine derivatives and of novel guanidinium-rich compounds derived from the biopolymer cyanophycin. Helv. Chim. Acta 101, e1800112 (2018).
Grogg, M., Hilvert, D., Beck, A. K. & Seebach, D. Syntheses of cyanophycin segments for investigations of cell-penetration. Synthesis 51, 31–39 (2019).
Acknowledgements
We thank members of the Schmeing laboratory for helpful advice and discussion, N. Rogerson for proofreading, staff at the McGill Facility of EM Research (M. Strauss, K. Basu and K. Sears) and APS (F. Murphy and S. Banarjee) for support during data collection. We thank the UCSD Cryo-Electron Microscopy Facility, which was supported in part by National Institutes of Health grants to T.S. Baker and a gift from the Agouron Institute to UCSD. This work was supported by a Canada Research Chair and NSERC Discovery grant no. 418420 to T.M.S., and by funding from the Swiss National Science Foundation and the ETH Zurich to D.H. We thank P. Emsley and R. Nicholls for help with restraints file generations. This work includes research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (grant no. P30 GM124165). The Eiger 16M detector on the 24-ID-E beamline is funded by a NIH-ORIP HEI grant (no. S10OD021527). This research used resources of the Advanced Photon Source, a US DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. Gene synthesis of TmCphA1 was conducted by the US DOE Joint Genome Institute, a DOE Office of Science User Facility, which is supported under contract no. DE-AC02-05CH11231, as part of JGI grant no. 503632 to T.M.S.
Author information
Authors and Affiliations
Contributions
T.M.S. and I.S. designed the study. I.S. performed all molecular biology and biochemical experiments. I.S., A.S.H., I.L. and T.M.S. performed cryo-EM data collection and initial structure determination. I.S. performed crystallography and structure determination, as well as modeling and refinement of all structures. M.G. performed chemical synthesis under the direction of D.S. and D.H. T.M.S. and I.S. wrote the paper and T.M.S., I.S., A.E.L., I.L. and D.H. edited it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemical Biology thanks Elke Dittmann, Satish Nair and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CphA1 distribution.
Phylogenetic tree of CphA1 sequences. A BLAST search found over 4000 CphA1-encoding gene sequences. Analysis of these sequences revealed that they are spread across most major bacterial phyla. Specific clades of particular interest were manually annotated and colored. The homologs used in this study are labeled in red. Gammaproteobacterial TmCphA1 and AbCphA1 share ~41% identity with cyanobacterial SuCphA1. There is evidence for both ancient horizontal gene transfer (alpha-, delta- and gammaproteobacterial CphA1s cluster together, but apart from betaproteobacteria1) and more recent transfer in the unlabeled, black clusters of CphA1s that are from several different bacterial groups.
Extended Data Fig. 2 CphA1 tetramerization.
a, SuCphA1 (a, c) and TmCphA1 (b, d) display different tetramer architectures, in which different monomers are responsible for tetramer-forming interactions. e, The EM map and structure of SuCphA1 showing the tetramer interface, which is centered on W672. f, Gel filtration chromatograms of all three CphA1 homologs used in this study, show they all form tetramers in solution.
Extended Data Fig. 3 Comparison of CphA1 active sites to homologous enzymes.
a, Overlay of SuCphA1 G domain and bifunctional glutathione synthetase from S. agalactiae (PDB code 3LN6) showing the similar ATP binding mode and conserved residues. b, Overlay of SuCphA1 G domain and glutathione synthetase from E. coli (PDB code 1GSA) showing the similar substrate orientation and overall structure. c, Overlay of SuCphA1 M domain and MurE ligase from M. tuberculosis residues (PDB code 2WTZ) showing the similar ATP binding mode and conserved, and (d) similar substrate orientation. e, The interactions made by cyanophycin with residues in the M domain of SuCphA1. f, The three versions of cyanophycin and cyanophycin analogs used for the determined structures of SuCphA1 presented in this study.
Extended Data Fig. 4 Flexibility, substrate binding and surface electrostatic potential of CphA1.
a, Local resolution estimates of the cryo-EM maps of tetrameric SuCphA1 (left) and dimeric AbCphA1 (right). b, Overlay of the two chains in the crystal structure of TmCphA1 (light blue) on the cryo-EM structure of SuCphA1 (colored), showing the different conformation adopted by Mlid of the crystal structure chain A and Glid in chain B. Mlid is not visible in chain B. c, Overlay of the unsharpened maps of SuCphA1 without cyanophycin substrate analogs (gray), and with (Asp-Arg)8-NH2 (red, right) and (Asp-Arg)8-Asn (blue, left). Clear extra density is visible in the maps calculated in the presence of substrate analogs, mostly near the active sites and the N domain. (Asp-Arg)8-Asn is also seen as product in the G domain active site, but no density is visible for the terminal Asn residue. d, Surface electrostatic potential maps of SuCphA1 and TmCphA1 dimers showing how the side that faces the active sites is lined with negatively and positively charged patched. The side facing the inner cavity, which is opposite the active sites, is mostly neutral. Active sites are marked with *, αa and αb are marked with rectangles.
Extended Data Fig. 5 CphA1 N domain structural homology, cyanophycin binding mode and mutants analysis.
a, Structure overlay of CphA1 with E. coli RNA polymerase alpha subunit (PDB code 4JK1), showing similarity in parts of their structures. b, Possible arrangement of cyanophycin that allows either its positive charges or negative charges to interact with αa or αb. c, Activity assays of TmCphA1 N domain mutants showing similar results to those observed for the equivalent SuCphA1 mutants (displayed in Fig. 5). n = 4 independent experiments. Data are presented as individual measurements and mean value, error bars represent SD values. d, Differential scanning fluorimetry melting curves and protein Tm values of CphA1 N domain mutants. The similarity of Tm values between wildtype and N domain mutants suggests that the observed differences in activity are a result of differences in interaction with cyanophycin rather than differences in protein stability. Additionally, the gel filtration profiles of the proteins during the purification process were all similar, again suggesting similar stability of wildtype and mutants. The values in the table represent the mean and SD of 3 independent measurements.
Extended Data Fig. 6 Dimeric CphA1 mutation schematics and activity in 100 mM sodium chloride.
a, The tetramerization interface, between W672 and residues 468–470, is disrupted in the obligate dimer W672A mutants. b, Cartoon representation of the CphA1 mutants. c, Both mutant combinations of dimer mutants (G+M+/G-M− and G+M−/G-M+) show the same decrease in activity level relative to wildtype. The ratio between the activity rate of the wildtype CphA1 and mutants is similar to that observed with no sodium chloride in the reaction buffer. n = 4 independent experiments. Data are presented as individual measurements and mean value, error bars represent SD values.
Extended Data Fig. 7 Fourier shell correlation for EM datasets.
FSC plots for all EM maps determined in this study.
Extended Data Fig. 8 Model of cyanophycin synthesis within wildtype and mutant CphA1.
Models of cyanophycin synthesis by WT CphA1 and the active site mutants used in the study.
Supplementary information
Supplementary Information
Supplementary Note, Fig. 1, Tables 1–6 and References.
Supplementary Video 1
The structure of SuCphA1. This video highlights the structure of the three domains, substrate binding interactions and tetramer interface.
Supplementary Video 2
Comparison of the structures of SuCphA1 and TmCphA1. This video focuses on the overall architectures of monomers, dimers and tetramers.
Supplementary Video 3
Four modes of 3D variability analysis of SuCphA1 bound with (β-Asp-Arg)8-Asn. This video shows the first mode from CryoSPARC2 3D variability analysis.
Supplementary Video 4
Model for the catalytic cycle of CphA1. This video shows the proposed model for biosynthesis of cyanophycin by SuCphA1.
Source data
Source Data Fig. 2
Statistical raw data Fig. 2a.
Source Data Fig. 2
Uncropped gel image Fig. 2b.
Source Data Fig. 3
Statistical raw data Fig. 3d.
Source Data Fig. 4
Statistical raw data Fig. 4d.
Source Data Fig. 5
Statistical raw data Fig. 5.
Source Data Fig. 6
Statistical raw data Fig. 6.
Source Data Extended Data Fig. 5
Statistical raw data Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Statistical raw data Extended Data Fig. 6.
Rights and permissions
About this article
Cite this article
Sharon, I., Haque, A.S., Grogg, M. et al. Structures and function of the amino acid polymerase cyanophycin synthetase. Nat Chem Biol 17, 1101–1110 (2021). https://doi.org/10.1038/s41589-021-00854-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00854-y
This article is cited by
-
Cyanophycin modifications for applications in tissue scaffolding
Applied Microbiology and Biotechnology (2024)
-
Recovery of cyanophycin granule polypeptide from activated sludge: carbon source dependence and aggregation-induced luminescence characteristics
Frontiers of Environmental Science & Engineering (2024)
-
Structural and functional insights into δ-poly-L-ornithine polymer biosynthesis from Acinetobacter baumannii
Communications Biology (2023)
-
Bioinformatics of cyanophycin metabolism genes and characterization of promiscuous isoaspartyl dipeptidases that catalyze the final step of cyanophycin degradation
Scientific Reports (2023)
-
A cryptic third active site in cyanophycin synthetase creates primers for polymerization
Nature Communications (2022)