Abstract
The genus Pseudoterranova includes parasite species of cetaceans and pinnipeds. The third stage larva (L3) of seal-infecting species occur in second intermediate or paratenic fish hosts mainly in neritic waters. This study firstly describes a Pseudoterranova L3 from meso/bathypelagic fishes off Macaronesia. L3s were morphologically and genetically studied by light microscopy and sequencing of the mtDNA cox2 and entire ITS rDNA genes. Bayesian inferences were performed with sequences from the larvae and selected sequences from GenBank. The nematode L3s were molecularly identified as Pseudoterranova ceticola, a parasite of kogiid whales. Such larvae were collected from Bolinichthys indicus, Chauliodus danae, Eupharynx pelecanoides, Diaphus rafinesquii, D. mollis, Diretmus argenteus and Maulisia argipalla. They mainly occurred in the viscera of these fishes. Pseudoterranova ceticola L3 were small (< 12 mm) and whitish, and a prominent characteristic is a circumoral ridge extending from the ventral boring tooth which differentiate them from Pseudoterranova spp. L3 maturing in pinnipeds and Terranova sensu lato larvae that mature in poikilotherms. The shape of the tail: conical, long, pointed, ventrally curved and lacking mucron also distinguish these larvae from those of the pinniped-infecting Pseudoterranova spp. Phylogenetic analyses based on mtDNA cox2 and ITS rDNA sequences suggest that P. ceticola is closely related to Skrjabinisakis spp., and not with Pseudoterranova spp. parasitizing pinnipeds. The related species Skrjabinisakis paggiae, S. brevispiculata and S. physeteris (until recently belonging to genus Anisakis), are as P. ceticola also parasites of physeteroid cetaceans. The morphology and morphological variation of the larvae of the cetacean parasite P. ceticola is thoroughly described for the first time. These L3 can readily be morphologically distinguished from those of the pinniped-infecting Pseudoterranova spp. The parasite likely completes its life cycle in the mesopelagic and bathypelagic realm, with meso/bathypelagic fish as 2nd intermediate or paratenic hosts and kogiids as final host. Thus, Pseudoterranova from cetaceans appear to be morphologically, genetically, and ecologically differentiated to those from pinnipeds, suggesting that they are not congeneric.
Similar content being viewed by others
Introduction
The taxonomy of ascaridoid nematodes remains confusing and unresolved. The issue is of particular importance since species from the genera Anisakis, Pseudoterranova and Contracaecum are recognized as causative agents of fish-borne zoonotic diseases of worldwide concern, i. e. anisakidoses1,2. Generally, these anisakids use crustaceans as first intermediate hosts, fish and squid as second intermediate or paratenic hosts, and marine mammals (i. e. cetaceans for Anisakis spp. and pinnipeds for Pseudoterranova spp. and Contracaecum spp.) as final hosts of their life cycle [reviewed by 3]. In addition, anisakid species belonging to the genus Terranova (which is now considered taxon inquirendum, see4 and further comments at the discussion section) englobed species parasites of elasmobranchs, teleosts, crocodilians, colubrid snakes and marine mammals4.
Identification of Terranova-like third-stage larvae (L3) present in fish intermediate or paratenic hosts is difficult. Larvae belonging to Pseudoterranova, Pulchrascaris and Terranova sensu lato are too morphologically similar to identify them even to genus5. The common morphological features are the presence of the excretory pore opening ventrally at the anterior end, presence of ventriculus without an appendix and having an intestinal caecum5,6,7,8. Molecular identification is therefore needed. Robust identification of anisakid parasites is crucial for understanding their distribution and epidemiology.
In the present study, a new Terranova-like larval type, collected from mesopelagic/bathypelagic fish species of Macaronesia, North West (NW) African waters, is morphologically described, and molecularly recognized as a potentially zoonotic member of the genus Pseudoterranova.
Methods
Fish collection
During May 2019, mesopelagic and bathypelagic fishes including the following host species: Bolinichthys indicus, Chauliodus danae, Eupharynx pelecanoides, Diaphus rafinesquii, Diaphus mollis, Diretmus argenteus and Maulisia argipalla were caught in waters off NW Africa from Cape Verde to North East (NE) of Madeira during a research cruise on board of the Norwegian vessel “RV Kronprins Haakon” (Table 1, Fig. 1). Hauls were conducted with 2 different gears: a macroplankton trawl (theoretical mouth opening 6 × 6 and 8 mm stretched mesh size) and a Multipelt trawl (mouth opening height of 35 m and 20 mm mesh in the cod-end). Fishes were frozen on board at − 20 °C for later parasite inspection on land. Fish samples were collected within the MEESO project (EU H2020 research and innovation programme, Grant Agreement No 817669) and procedures were carried out in accordance with the relevant EU legislation including EU Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Norwegian research vessels have authorization to collect fishes for research purposes; in addition, permission for the collection of the present fishes was obtained from coastal countries.
Trawl stations from which infected fish hosts were obtained. Positions: station (st.) 4601: 17.969 N, 23.956 W; st. 4604: 26.899 N, 19.232 W; st. 4606: 29.767 N, 16.087 W; st. 4610: 33.695 N, 13.232 W. Figure 1 was created using R version 4.95 (2021–03-31) (https://www.r-project.org/) implemented in RStudio 1.4.1106 (https://www.rstudio.com/) using.
Parasite collection
Fishes were thawed at room temperature, opened and the visceral organs and emptied body cavity were placed in a petri dish with physiological saline solution and examined under stereomicroscope for ascaridoid nematodes. The parasites were collected, and the internal organs and carcass were then placed into plastic bags and refrozen. These were later examined using the UV-press method9, to detect any larvae not recovered during dissection, specially from the musculature.
Morphological study
The nematode larvae were examined in temporary mounts in physiological saline solution, and photographed. Various morphotypes were recognized, but only findings concerning Terranova-like larvae8,10 (N = 35) will be presented here. In addition, infection levels such as parasite prevalence and abundance will be published elsewhere.
Morphometric measurements
Series of digital photographs were obtained from 33 larvae. Measurements were taken from the digital images, except larval body lengths that mostly were measured at a mm scale.
Measurements from images were obtained using the software Image J (https://imagej.nih.gov/ij/). The oesophagus, ventricle and tail lengths were taken along the midline. The caecum length was measured from the aperture into the ventricle to the caecum end (Fig. 2).
Measurements taken from the images of Terranova-like larvae. The oesophagus length is taken along the midline from the start of the oesophagus (i. e. slightly sub-terminally in the roundworm) to the ventricle. The caecum length was measured from the aperture into the ventricle to the caecum end. The tail length represents the distance along the midline, from the level of the anus/cloaca to the posterior end. Figure 2 was created in Adobe Photoshop 23.5.0 (https://www.adobe.com/photoshop).
Molecular analyses
DNA was extracted from a randomly selected subsample of 19 nematode larvae using the DNeasy Blood & Tissue Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions with the modification that sample lysis was enhanced by mechanical disruption with ceramic bead-beating system (Precellys ceramic kit 2.8 MM, VWR and Precellys 24 Tissue Homogenizer, Bertin Technologies). DNA was eluted with 30 µl AE buffer.
Polymerase chain reaction (PCR) were done with primers that amplify the entire internal transcribed spacer (ITS) region of the nuclear ribosomal DNA (ITS1-5.8SrRNA -ITS2). The NC5F (5’–GTAGGTGAACCTGCGGAAGGATCATT- 3’) and NC2R (5’ TTAGTTTCTTTTCCTCCGCT -3’) primers were used. PCR conditions followed Zhu et al.11, but annealing temperature was 54 °C instead of 55 °C.
The mitochondrial cytochrome c oxidase subunit II (cox2) gene was amplified using the primers 211F (5′-TTTTCTAGTTATATAGATTGRTTTYAT-3′) and 210R (5′-CACCAACTCTTAAAATTATC-3′)12 according to Mattiucci et al.13 with the following modifications. The cycling conditions were an initial denaturation at 94 °C for 5 min, followed by 35 cycles of: denaturation at 94 °C for 30 s, annealing at 46 °C for 1 min, extension at 72 °C for 90 s; followed by final step of final extension at 72 °C for 10 min, and hold at 4 °C.
PCR products were sent for purification and sequencing (using the primers NC5F and 210R) to Eurofins (Cologne, Germany). The National Center for Biotechnology Information (NCBI) sequence database (henceforth ‘GenBank’) was searched for similar sequences using BLAST (Basic Local Alignment Search Tool) (USA)14.
Phylogenetic analyses
Sequences generated in this study were aligned with selected sequences obtained from GenBank, using CLUSTAL W in MEGA X (Table 2 and 3)15. High similarity scores in the BLAST as well as larvae morphological similarity were used as the criteria to select the sequences. The default setting parameters of ClustalW were used, and the alignments were manually edited and trimmed in MEGA X. Toxocara canis and Ascaris lumbricoides were set as outgroup for the cox2 phylogenetic analysis. Due to indel-induced alignment problems in ITS16, only the closely related Anisakis spp., and Pseudoterranova spp. could be aligned with confidence in homology. For the same reason, no outgroup was included. The entire ITS sequences of Pseudoterranova krabbei and Pseudoterranova bulbosa identified from two cod (Gadus morhua) caught in northern Norwegian waters were sequenced and used in the analysis (Table 2).
Phylogenetic analyses were inferred using Bayesian inference (BI) method in BEAST v1.10.417. The optimum evolutionary models were estimated using the Bayesian information criterion (BIC) as implemented in MEGA X. The optimum model was K2 + G for the ITS dataset based on BIC, independently of using all sites or complete deletion (i. e. gaps/missing data treatment). However, this model was not available in BEAST, so we used the best next best model available, i.e. HKY + G for the BI. The optimum model was HKY + G + I for cox2 dataset based on BIC criteria. The BEAST file was previously generated in BEAUti with the following characteristics: sites: entering the best substitution model and otherwise default settings; clock type: strict clock; tree prior: Speciation: Yule process; MCMC: length of chain = 10,000,000, echo state to screen every = 1000, log parameters every = 1000. Effective sample size of parameters (i. e. > 200) was checked in Tracer v.1.7.218. The created tree was drawn in TreeAnnotator v1.10.4 and the burnin as the number of states was specified at 100,000. Figtree v1.4.4 was used to visualize the phylogenetic trees.
Results
A total of 35 Terranova-type larvae were recovered. All these were morphologically similar. They were found in 7 fish species (Table 1). In-situ, the larvae were coiled like a coil or watch spring. Most (94%) were found in the viscera, but two larvae were found in the muscle of D. rafinesquii. The larvae had a light neon-bluish colour when exposed to UV-light, after freezing.
Morphology
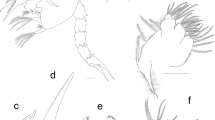
Larvae were small and pale, with a thick-set appearance (Fig. 3, Table 4). The body was widest at the middle and posteriorly. Body length: max width ratio was 22—36:1 (mean 28 ± 4, N = 27). The cuticle was smooth, but with inconspicuous transverse striae which were most evident in tail. At anterior end, lip anlagen were visible through the cuticle, and associated with surface bulbs (Fig. 3c). Prominent conical boring tooth at the anterior extremity between ventro-lateral lip anlagen, projecting anteroventral at an angle of about 130° (115°–145°) to main axis. The boring tooth base gives rise to a circumoral cuticular ridge (Fig. 3b). The dorsoventral extent of this ridge from the boring tooth tip was 40.8 ± 3.2 µm (mean ± SD) (range = 36–49 µm; N = 25). The excretory pore was ventrally located, near the base of the boring tooth. The oesophagus was clavate, widest posterior. The nerve ring was positioned within the anterior 25 (20–27) % (mean (range); N = 12) of the oesophagus length. The oesophagus length constituted 10 (9–13) % (mean (range)) of the body length. The ventricle was prominent, wider than the posterior oesophagus; oesophagus to ventricle length ratio was 1.5–2.6:1 (2.0 ± 0.2; N = 27). The caecum was normally shorter than the ventricle, averaging 74% of its length (range = 50–119, SD = 18). The tail was elongate conical, curved ventrally, pointed and without a distinct mucron (Fig. 3–G).
Digital photographs of P. ceticola L3 from Macaronesian deepwater fishes. (a) Entire P. ceticola larva from Diaphus mollis. (b) Anterior end showing the ventral boring tooth connected to the cuticular ridge, arrow indicates the excretory pore. (c) out-of-focus view of the anterior end, showing bulbs (arrows). (d) Anterior end showing nerve ring (NR). (e–f) ventricle region, showing ventricle (V) and caecum (C), intestine (Int), and oesophagus (Oe). Arrow in (f) indicates aperture intestine-ventricle. (g) Tail region, arrow indicating anus. (h) Part of tail, showing transverse cuticular striation. Specimens were collected from D. mollis (a,b,d,f,g) and D. rafinesquii (c,e,h). Scale bars: (a) 500 µm, (b, c) to same scale 50 µm, (d) 100 µm, (e, f) to same scale 200 µm, (g) 100 µm, (h) 50 µm.
Molecular identification
The ITS sequences (801–842 bp) obtained from 13 Terranova type larvae were 100% identical. However, ambiguous positions (i. e. double signals) were seen in the sequences from five of these worms. The cox2 sequences (570–580 bp) obtained from 18 Terranova type larvae showed 97.1–99.4% identity. With a single exception (in DiRa38), all substitutions were silent. The cox2 sequences were 96.9% to 97.9% similar to a Pseudoterranova ceticola cox2 sequence from a Caribbean Sea K. breviceps (GenBank accession number DQ116435). Blast searches with the ITS sequence revealed 99.6–100% identity to sequences from adult worms (found in kogiid whales) or larvae (from marine fish and agnathans) identified as Anisakis sp. (see Supplementary file: Table S1, and further comments at discussion section). In addition, the ITS sequence of a P. ceticola larva from the Tanzanian fish Saurida undosquamis (ON128286)19 was 100% identical. Sequences of the presently identified P. ceticola L3 were deposited in GenBank with the accession numbers (ITS: OP352234- OP352246) and (cox2: OP380493-OP380510) (see also Supplementary file: Table S2).
Phylogenetic analyses
Phylogenetic analyses were performed on ITS rDNA and mt DNA cox2 datasets.
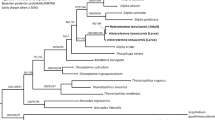
In the cox2 BI tree, adult P. ceticola from K. breviceps (DQ116435), larva from the fish S. undosquamis (ON155434), larva from the fish Katsuwonus pelamis (LC712860) and the sequences of the present Terranova-like larvae group together in a well-supported clade, representing a sister group to a clade with Skrjabinisakis physeteris, S. brevispiculata and S. paggiae (until recently belonging to the genus Anisakis (see20,21) and related sequences (Fig. 4 and see also Figure S1 at Supplementary files which details the intraspecific variations and relationships of P. ceticola). The major clade (Clade A) with these two subclades is a well-supported sister group to a clade containing the A. simplex complex (A. simplex (s.s.)), A. pegreffii, A. berlandi), Anisakis typica, Anisakis nascettii, Anisakis ziphidarum and Pseudoterranova spp. from pinnipeds (P. azarasi, P. bulbosa, P. cattani, P. decipiens (s .s.) and P. krabbei) (Clade B).
Phylogenetic tree from Bayesian inference based on cox2 sequences. A: clade A; B: clade B. Figure 4 was created in Figtree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
The unrooted tree obtained based on the ITS region sequences also supported Clade A and its two subclades (Fig. 5). Again, the pinniped Pseudoterranova spp. grouped separately. Also, the sequences of the present Terranova-like larvae grouped with larval and adult genotypes of worms identified as belonging to genus Anisakis, from fish and kogiid whales. These included worms from an Australian K. breviceps and a Philippine K. sima, a larva from the teleost fish Pagellus bogaraveo from Madeira and from the fish Epinephelus areolatus from Indonesia.
Unrooted tree from Bayesian inference based on ITS sequences. Figure 5 was created in Figtree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Discussion
Genus Terranova was erected for a New Zealand shark parasite, T. antarctica22,23. Later additions of Terranova spp. represent further parasites from elasmobranchs, but also parasites from teleosts, crocodilians, colubrid snakes and marine mammals (reviewed by4). Attempts have been made to split the genus, and now the best-known species from poikilotherms (i. e. elasmobranchs, teleosts and reptiles) are allocated to genera Euterranova, Neoterranova or Pulchrascaris4. Several lesser known species are retained in Terranova sensu lato (species inquirenda, see4). Genus Phocanema was proposed for Porrocaecum decipiens24, a Terranova-like pinniped parasite that subsequently was shown to represent several cryptic species (see3,25). A new genus, Pseudoterranova26, was proposed for Terranova kogiae from an Australian kogiid whale Kogia breviceps27, based on an erroneous interpretation of the excretory pore position28. Gibson28 corrected this error, but retained and redefined Pseudoterranova on the basis of the anterior extent of the glandular left filament of the excretory system. This genus now contains all the Terranova-like species from homeotherms, P. kogiae and P. ceticola from whales, and several species from pinnipeds, with Phocanema as a synonym.
The presently described nematode larvae were all morphologically similar, thus resembling a single morphotype. They show morphological characters shared by genera Terranova sensu lato, Pseudoterranova and Pulchrascaris, such as an excretory pore at the base of the ventral lip and the presence of an intestinal caecum. Recently, Terranova sp. type 1 and 2 larvae sensu Cannon8 from teleost paratenic hosts have been molecularly found to include Pulchrascaris australis and Terranova pectinolabiata (now Euterranova pectinolabiata) from Australian shark definitive hosts, respectively4,8,29,30. Gonzalez-Solis et al.31 used morphology alone to identify Terranova type 1 larvae from Hawaiian fishes as Pulchrascaris sp. In general, larvae of Pulchrascaris spp. and Terranova spp. that are present in teleost paratenic hosts and mature in elasmobranchs, show very long and slender ventricles, alongside an even longer caecum, and they have conical pointed tails without mucron5,8,31,32. The Terranova-like larvae described in the present study however have shorter more oval ventricles, accompanied by caeca that rarely exceed the ventricle in length. These characteristics are shared with larvae of Pseudoterranova spp. from seals and sea lions. However, the tails of the Pseudoterranova spp. larvae of pinniped infecting species differ from those of P. ceticola in being generally short and rounded with a mucron10,33,34,35. In addition, the boring tooth inclines ventrally and is prominent, different to those present in Pseudoterranova spp. from pinnipeds which are straighter and comparatively smaller35,36,37,38. The larvae presented in this study also appear to be considerably smaller in size, reaching only up to 12 mm in body length, compared to P. decipiens s.l. (10–60 mm), P. cattani (17–43 mm) or P. decipiens sp. E (20–38 mm)6,33,34,35,36,38,39. A most prominent character that distinguish the present larvae from pinniped Pseudoterranova spp. is the circumoral cuticular ridge connected with the boring tooth. This character also distinguishes them from most Terranova sensu lato larvae, that may represent elasmobranch parasites. However, Deardorff et al. found a Terranova larval type in Hawaiian teleosts, designated Terranova sp. type HA40. Those larvae fit the larvae presented in here in most aspects including caecum:ventricle length ratio, tail shape and the presence of an anterior circumoral ridge (Table 5). The Terranova sp. type HA larvae were found to survive in and penetrate the stomach wall of rats, demonstrating a zoonotic potential40,41. This suggests that they mature in a homeotherm, i. e., belong in genus Pseudoterranova. Similarly, Kuramochi et al. described Pseudoterranova cf. ceticola larvae recovered from stranded Japanese K. breviceps and the Risso’s dolphin (Grampus griseus) (see Table 5)42, and those larvae fit in most aspects the present larvae and the Terranova sp. type HA of Deardorff et al.40, thus suggesting to be conspecific. Most recently, a single P. ceticola larva from the scombrid fish K. pelamis in Japan was briefly described 21 (see Table 5). The larva, even though smaller, resembles in most aspects to our larva, however, authors did not refer to the circumoral cuticular ridge, which is apparent in their Fig. 1A 21, as the most prominent character of P. ceticola L3.
The present larvae are molecularly identified as Pseudoterranova ceticola, a parasite of kogiid whales. That finding suggests that the Terranova sp. type HA larvae are also likely P. ceticola larvae. Kogia spp. are common around the Hawaii islands (see Bloodworth and Odell46). Pseudoterranova ceticola was originally described from Kogia sima stranded in Mississippi, USA47. Another species, P. kogiae was described from K. breviceps in Australia. Pseudoterranova kogiae differs from P. ceticola by having more pairs of caudal papillae present in the males and by having three transverse rows of plectanes which are absent in P. ceticola27,47,48. These species appear to show a caecum equal or longer than the ventricle, even in juvenile specimens of P. ceticola reported from the final hosts (Table 5)27,42,44,47. Therefore, the relationship between caecum length and ventricle length in larvae from intermediate or transport hosts may not reflect the adult morphology, contrary to the arguments of González Solís et al.31. In addition, these species from kogiid whales are small compared to adult males maturing in pinnipeds. Pseudoterranova ceticola and P. kogiae ranged between 12 and 26 mm and 20–30 mm long, respectively, whereas the mean ± SD (range) for P. decipiens s.s., P. krabbei, P. bulbosa, P. cattani and P. azarasi were reported as 44 ± 7 and 48 (42–54), 36 ± 3 and 35 (31–43), 47 ± 5 and 48 ± 7, 40 ± 9 (26–62) and 49 ± 3 mm long, respectively27,42,44,47,49,50,51,52.
The present identification relies on the high identity of the cox2 sequences with the reference sequence of P. ceticola collected from a Caribbean Sea K. breviceps53. Similarly, several P. ceticola collected from K. breviceps and K. sima stranded in Puerto Rico and Florida (USA) were morphologically and/or molecularly identified by sequencing analysis of the mtDNA cox254,55, so the association of this genotype with P. ceticola should not be in doubt. However, the herein presented ITS1-5.8S-ITS2 sequences from the same P. ceticola larvae specimens showed 100% identity to sequences from some worms identified as Anisakis sp.56,57,58,59,60 (see Table S1 at supplementary file). It is probable that this occurred due to the lack/impossibility of a morphological examination (probably hampered by a poor condition of worms in those studies) revealing the characteristic caecum of P. ceticola, in combination with a GenBank match for Anisakis sp. In relation to this, in cleared specimens the caecum may become very pale and not easy to distinguish, hence it might be overlooked (per. obs., see also61). The diagnostic RFLP pattern of P. ceticola with the HhaI restriction enzyme produce 4 bands, of about 80, 180, 200 and 400 bp length, and a single undigested band of about 1000 bp with HinfI55,62. However, this pattern has also been obtained with worms that may have been erroneously identified as Anisakis sp.57,58,59,63,64, leading to confusion. A list of the anisakids likely representing P. ceticola but identified as Anisakis sp. is provided in the Supplementary file Table S3.
Clearly, one reason for this confusion is the lack of a morphological account of P. ceticola larvae, which we provide here. However, trustable sequence information from P. kogiae is not available. Either P. kogiae and P. ceticola are two valid species, or they are synonymous. Gibson examined the types of P. kogiae and also had at hand specimens of P. ceticola from South African K. breviceps, and seems to consider them valid species28. Shamsi et al.65 provided an ITS region sequence (MK325217) of an “Anisakis sp.” from an Australian K. breviceps, interestingly being the same type host and geographical area where P. kogiae was described27. The sequence was considerably different to our P. ceticola sequences by having a unique 3 bp insert plus 2 substitutions (see also Fig. 5). Clearly, more research is necessary in order to clarify the relationship between these Pseudoterranova spp.
Phylogenetic analysis obtained from the mtDNA cox2 sequences, suggests that the clade with P. ceticola sequences is closely related to a clade formed by S. paggiae, S. cf. paggiae, S. brevispiculata, S. physeteris and Skrjabinisakis sp. 2 (species which until most recently belonged to the genus Anisakis20,21). The BI analysis of the ITS region also supports that P. ceticola is closely related to the former S. physeteris complex clade. These results are similar to those by Cavallero et al.28 and most recently to those by Takano & Sata21. The monophyly of the clade formed by P. ceticola and S. paggiae, S. brevispiculata and S. physeteris was suggested by Cavallero et al. based on phylogenetic analyses of the cox2 and ITS regions, as found here. However, Takano & Sata found the species Neoterranova caballeroi from reptiles as the most related species to P. ceticola which raises concerns21. We have performed an additional BI analysis including the cox2 sequence of N. caballeroi (AF179921), and N. caballeroi was placed as an offshoot of Anisakinae species (i. e. Anisakis, Skrjabinisakis and Pseudoterranova), which we believe is more congruent with the ecology and morphology of this parasite (see figure S2 at Supplementary files).
In this study, P. ceticola was collected from fishes which distributions span the meso- and bathypelagic zones, caught off Cape Verde, Canary Islands and Madeira. Pseudoterranova ceticola larva in the deep-water shark Centrophorus squamosus taken off Madeira was also reported by Costa et al.62, who identified it by ITS-RFLP but provided no sequence information. Recently, Cipriani et al. identified a single P. ceticola larva from the reef-associated fish S. undosquamis caught between 100–600 m depth off the coast of Tanzania, by sequencing analysis of the ITS rDNA and mtDNA cox2 sequences19. Most recently, a single P. ceticola larva from the scombrid fish K. pelamis was also identified in Japan21. It appears possible that the former fish species, i. e. C. squamosus, S. undosquamis or K. pelamis, could have acquired P. ceticola though predation upon parasitized meso/bathypelagic fish.
It appears then that P. ceticola may have different host specificity depending on life stages, being a host specialist in the final host (i. e. kogiid whales), and generalist in the second intermediate or paratenic host (i. e. fishes). Adult P. ceticola has only been found in kogiid whales (i.e. K. sima and K. breviceps) suggesting stenoxeny at the final host level. Pseudoterranova ceticola was reported in K. sima from the Gulf of Mexico, Japan, Caribbean and SE Atlantic coasts of USA, and from K. breviceps in the same geographical region (presumably only as larvae in Japan), Atlantic Canada, NW Spain and South Africa28,42,44,47,48,53,54,55,66,67,68. In addition, Pseudoterranova sp. (as Terranova sp.) has been reported in K. breviceps from Brazil, Pacific Gulf of California (Mexico) and France, and in K. breviceps and K. sima from the Caribbean region45,68,69,70. Pseudoterranova cf. ceticola L3 has been reported from two Japanese G. griseus and Pseudoterranova sp. has been reported from Caribbean pygmy killer whale (Feresa attenuata)42,68.
Thus, P. ceticola has so far been found in temperate waters of western and eastern Atlantic and Pacific Oceans (see above and Table S3), a distribution apparently overlapping that of its final kogiid hosts71.In addition, kogiids have been reported stranded or observed in Macaronesia areas46,71,72,73. Contrarily, Pseudoterranova spp. from pinnipeds are mainly distributed in Boreal and Austral waters where their final hosts thrive74.
Our results suggest that the life cycle of P. ceticola occur in the mesopelagic and bathypelagic realm. In addition, the parasite also appears to occur in benthopelagic, demersal and even reef-associated fish hosts (see Table S3). Contrarily, primarily neritic, benthic and demersal fishes appears to be involved in transmitting Pseudoterranova spp. to pinnipeds3,6,36,74,75,76,77. Indeed, it has been observed that P. decipiens s.l. larvae hatched from eggs adhered to the substrate by their tails76,78, being eaten by benthic meiofauna (mainly copepods) first intermediate hosts leading to transmission up a benthic food-web76,79.
There are some studies which have indicated that meso- and bathypelagic fish are prey for K. breviceps and K. sima46,72,80. West et al.80 analysed the stomach content of stranded K. breviceps of the Hawaiian archipelago and identified among others D. argenteus, Diaphus sp. and E. pelecanoides, which are species that were found infected by P. ceticola in the present study. The parasite might also be transmitted through the food web from mesopelagic fish to squids (and other fishes) and then to the whales, since mid and deep-water cephalopods are also known as a very important part of the diet of these kogiids71,72,80.
Conclusions
Pseudoterranova ceticola third-stage larva (L3) was herein fully described for the first time. The parasite was recovered from meso- and bathypelagic fishes from off Macaronesia archipelagos (NW Africa). L3 were small, pale, with a thick-set appearance and bluish when exposed to UV-light after thawing. Ventricle morphology, presence of a caecum, tail shape and the presence of a circumoral cuticular ridge extending dorsally from boring tooth are morphological characteristics that aid identification. Pseudoterranova ceticola, which has kogiid (Physeteroidea) whales as final hosts, is related to Skrjabinisakis spp. (whose species formerly belonged to the genus Anisakis) maturing in physeteroid whales, rather than to Pseudoterranova spp. from pinnipeds. This is evidence that genus Pseudoterranova may have to be split.
Data availability
Data supporting the conclusions of this article are included within the article and its supplementary files. The DNA sequences of the P. ceticola specimens identified were deposited in the public sequence repository GenBank (NCBI National Center for Biotechnology Information—https://www.ncbi.nlm.nih.gov/genbank), and their accession numbers can be found in the present manuscript (Table 2 and 3) and/or supplementary files (Table S2).
References
Buchmann, K. & Mehrdana, F. Effects of anisakid nematodes Anisakis simplex (s.l.), Pseudoterranova decipiens (s.l.) and Contracaecum osculatum (s.l.) on fish and consumer health. Food Waterborne Parasitol. 4, 13–22. https://doi.org/10.1016/j.fawpar.2016.07.003 (2016).
Mattiucci, S., Cipriani, P., Levsen, A., Paoletti, M. & Nascetti, G. Molecular epidemiology of Anisakis and anisakiasis: An ecological and evolutionary road map. Adv. Parasitol. https://doi.org/10.1016/bs.apar.2017.12.001 (2018).
Mattiucci, S., Cipriani, P., Paoletti, M., Levsen, A. & Nascetti, G. Reviewing biodiversity and epidemiological aspects of anisakid nematodes from the North-east Atlantic Ocean. J. Helminthol. https://doi.org/10.1017/S0022149X1700027X (2017).
Moravec, F. & Justine, J.-L. Erection of Euterranova n. gen. and Neoterranova n. gen. (Nematoda, Anisakidae), with the description of E. dentiduplicata n. sp. and new records of two other anisakid nematodes from sharks off New Caledonia. Parasite 27, 58. https://doi.org/10.1051/parasite/2020053 (2020).
Shamsi, S. & Suthar, J. Occurrence of Terranova larval types (nematoda: Anisakidae) in Australian marine fish with comments on their specific identities. Peer J. 4, e1722. https://doi.org/10.7717/peerj.1722 (2016).
Timi, J. T. et al. Molecular identification, morphological characterization and new insights into the ecology of larval Pseudoterranova cattani in fishes from the Argentine coast with its differentiation from the Antarctic species, P. decipiens sp. E (Nematoda: Anisakidae). Vet. Parasitol. 199, 59–72 (2014).
Deardorff, T. L. Redescription of Pulchrascaris chiloscyllii (Johnston and Mawson, 1951) (Nematoda: Anisakidae), with comments on species in Pulchrascaris and Terranova. Proc. Helminthol. Soc. Wash. 54, 28–39 (1987).
Cannon, L. R. G. Some larval ascaridoids from south-eastern queensland marine fishes. Int. J. Parasitol. 7, 233–243 (1977).
Levsen, A. & Lunestad, B. T. Anisakis simplex third stage larvae in Norwegian spring spawning herring (Clupea harengus L.), with emphasis on larval distribution in the flesh. Vet. Parasitol. 171, 247–253 (2010).
Berland, B., (1989) Identification of fish larval nematodes from fish. In: Möller H, editor. Nematode problems in North Atlantic fish. Report from a workshop in Kiel, 3 4 16–22.
Zhu, X., D’Amelio, S., Paggi, L. & Gasser, R. B. Assessing sequence variation in the internal transcribed spacers of ribosomal DNA within and among members of the Contracaecum osculatum complex (nematoda: Ascaridoidea: Anisakidae). Parasitol. Res. 86, 677–683 (2000).
Nadler, S. A. & Hudspeth, D. S. S. Phylogeny of the ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology hypotheses of structural and sequence evolution. J. Parasitol. 86, 380–393. https://doi.org/10.1645/0022-3395(2000)086[0380:POTANA]2.0.CO;2 (2000).
Mattiucci, S. et al. Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi n. sp. for A simplex sp. C (Nematoda: Anisakidae). J. Parasitol. 100, 199–214. https://doi.org/10.1645/12-120.1 (2014).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Nagy, L. G. et al. Re-mind the gap! Insertion - deletion data reveal neglected phylogenetic potential of the nuclear ribosomal internal transcribed spacer (ITS) of fungi. PLoS ONE 7, 1–9 (2012).
Suchard, M. A. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, 1–5 (2018).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Cipriani, P. et al. Anisakid nematodes in Trichiurus lepturus and Saurida undosquamis (Teleostea) from the South-West Indian Ocean : Genetic evidence for the existence of sister species within Anisakis typica (s.l.), and food-safety considerations. Food Waterborne Parasitol. 28, e00177. https://doi.org/10.1016/j.fawpar.2022.e00177 (2022).
Safonova, A. E. First report on molecular identification of Anisakis simplex in Oncorhynchus nerka from the fish market, with taxonomical issues within Anisakidae. J. Nematol. 53(1), 10. https://doi.org/10.21307/jofnem-2021-023 (2021).
Takano, T. & Sata, N. Multigene phylogenetic analysis reveals non-monophyly of Anisakis s.l. and Pseudoterranova (Nematoda: Anisakidae). Parasitol. Int. 91, 102631. https://doi.org/10.1016/j.parint.2022.102631 (2022).
Leiper, R. T. & Atkinson, E. L. Parasitic worms, with a note on a free-living nematode. British Museum (Natural History). Bristish Antarctic (“Terra Nova”) expedition, 1910. Natural History Report. Zool 2(3), 19–60 (1915).
Leiper, R. T. & Atkinson, E. L. Helminthes of the British Antarctic expedition 1910–1913. Proc. Zool. Soc. London, 222–226 (1914).
Myers, B. J. Phocanema, a new genus for the anisakid nematode of seals. Can. J. Zool. 37, 459–465 (1959).
Mattiucci, S., Paoletti, M., Webb, S. C. & Nascetti, G. Pseudoterranova and Contracaecum. In Molecular detection of human parasitic pathogens (ed. Liu, D.) 645–656 (CRC Press, 2012).
Mozgovoĭ, A.A., (1953) Ascaridata of animals and man, and the diseases caused by them. In: Osnovy nematodologii. Vol. II. Izd. AN SSSR, Moskva (In Russian)
Johnston, T.H., Mawson, P.M., (1939) Internal parasites of the pigmy sperm whale. Rec. South Aust Museum.6. http://www.biodiversitylibrary.org/item/126147.
Gibson, D. I. The systematics of ascaridoid nematodes-a current assessment. In Stone A (eds Platt, H. & Khalil, L.) 321–338 (Academic Press, 1983).
Shamsi, S., Barton, D. P. & Zhu, X. Description and characterisation of Terranova pectinolabiata n. sp. (Nematoda: Anisakidae) in great hammerhead shark, Sphyrna mokarran (Rüppell, 1837), in Australia. Parasitol. Res. 118, 2159–2168. https://doi.org/10.1007/s00436-019-06360-4 (2019).
Shamsi, S., Barton, D. P. & Zhu, X. Description and genetic characterisation of Pulchrascaris australis n. sp. in the scalloped hammerhead shark, Sphyrna lewini (Griffin & Smith) in Australian waters. Parasitol. Res. https://doi.org/10.1007/s00436-020-06672-w (2020).
González-Solís, D. et al. Parasitic nematodes of marine fishes from Palmyra Atoll, East Indo-Pacific, including a new species of Spinitectus (Nematoda, Cystidicolidae). Zookeys. 2019, 1–26 (2019).
Jabbar, A. et al. Larval anisakid nematodes in teleost fishes from Lizard Island, northern great barrier reef Australia. Mar. Freshw. Res. 63, 1283. https://doi.org/10.1071/MF12211 (2012).
ICES. (2012) Pseudoterranova larvae (“codworm”; Nematoda) in fish. Revised and updated by Matt Longshaw. ICES Identification Leaflets for diseases and parasites of fish and shellfish. Leaflet No. 7. 4 pp.
Arai, H. P. & Smith, J. W. Guide to the parasites of fishes of Canada part V: Nematoda. Zootaxa 4185, 1. https://doi.org/10.11646/zootaxa.4185.1.1 (2016).
Hurst, H. J. Identification and description of larval Anisakis simplex and Pseudoterranova decipiens (anisakidae: Nematoda) from New Zealand waters. New Zeal J. Mar. Freshw. Res. 18, 177–186 (1984).
Hernández-Orts, J. S. et al. Description, microhabitat selection and infection patterns of sealworm larvae (Pseudoterranova decipiens species complex, Nematoda: Ascaridoidea) in fishes from Patagonia Argentina. Parasite Vector. 6, 1–15 (2013).
Shiraki, T. Larval nematodes of family anisakidae (Nematoda) in the northern sea of Japan as a causative agent of eosinophilic phlegmone of granuloma in the human gastro-intestinal tract. Acta Med. Biol. 22, 57–98 (1974).
Berland, B. Nematodes from some Norwegian marine fishes. Sarsia 2, 1–50. https://doi.org/10.1080/00364827.1961.10410245 (1961).
George-Nascimento, M. & Llanos, A. Micro-evolutionary implications of allozymic and morphometric variations in sealworms Pseudoterranova sp. (Ascaridoidea: Anisakidae) among sympatric hosts from the Southeastern Pacific Ocean. Int. J. Parasitol. 25, 1163–1171 (1995).
Deardorff, T. L., Kliks, M. M., Rosenfeld, M. E., Rychlinski, R. A. & Desowitz, R. S. Larval, ascaridoid nematodes from fishes near the Hawaiian Islands, with commonents on pathogenicity experiments. Pacific Sci. 36, 187–201 (1982).
Deardorff, T. L., Kliks, M. M. & Desowitz, R. S. Histopathology induced by larval Terranova (Type HA) (nematoda: Anisakinae) in experimentally infected rats. J. Parasitol. 69, 191–195 (1983).
Kuramochi, T. et al. Stomach nematodes of the family anisakidae collected from the cetaceans stranded on or incidentally caught off the coasts of the Kanto districts and adjoining areas. Mem. Nat. Museum. Nat. Sci. 37, 177–192 (2001).
Deardorff, T. L., Raybourne, R. B. & Desowitz, R. S. Description of a third-stage larva, Terranova type Hawaii A (nematoda: Anisakinae), from Hawaiian fishes. J. Parasitol. 70, 829–831 (1984).
González-Solís, D., Vidal-Martínez, V. M., Antochiw-Alonso, D. M. & Ortega-Argueta, A. Anisakid nematodes from stranded pygmy sperm whales, Kogia breviceps (Kogiidae), in three localities of the Yucatan peninsula. Mexico. J. Parasitol. 92, 1120–1122 (2006).
Santos, C. P. & Lodi, L. Occurrence of Anisakis physeteris Baylis, 1923 and Pseudoterranova sp. (Nematoda) in pygmy sperm whale Kogia breviceps (De Blainvillei, 1838) (Physeteridae) in northeastern coast of Brazil. Mem. Inst. Oswaldo Cruz. 93, 187–188. https://doi.org/10.1590/s0074-02761998000200009 (1998).
Bloodworth, B. E. & Odell, D. K. Kogia breviceps (cetacea: Kogiidae). Mam. Species. 819, 1–12. https://doi.org/10.1644/819.1 (2008).
Deardorff, T. L. & Overstreet, R. M. Terranova ceticola n. sp. (Nematoda: Anisakidae) from the dwarf sperm whale; Kogia simus (Owen), in the Gulf of Mexico. Syst. Parasitol. 3, 25–28 (1981).
Abollo, E., Santiago, P., (2002) SEM study of Anisakis brevispiculata Dollfus, 1966 and Pseudoterranova ceticola (Deardoff and Overstreet, 1981) (Nematoda: Anisakidae), parasites of the pygmy sperm whale Kogia breviceps. Sci. Mar. 66 3 49 255
Di Deco, M. A., Orecchia, P., Paggi, L. & Petrarca, V. Morphometric stepwise discriminant analysis of three genetically identified species within Pseudoterranova decipiens (Krabbe, 1878) (Nematoda: Ascaridida). Syst. Parasitol. 29, 81–88 (1994).
George-Nascimento, M. & Urrutia, X. Pseudoterranova cattani sp. nov. (Ascaridoidea: Anisakidae), a parasite of the South American sea lion Otaria byronia De Blainville from Chile. Rev. Chil. Hist. Nat. 73, 93–98. https://doi.org/10.4067/s0716-078x2000000100010 (2000).
Mattiucci, S. et al. Allozyme and morphological identification of Anisakis, Contracaecum and Pseudoterranova from Japanese waters (Nematoda, Ascaridoidea). Syst Parasitol. 40, 81–92 (1998).
Paggi, L. et al. Pseudoterranova decipiens species A and B (Nematoda, Ascaridoidea): Nomenclatural designation, morphological diagnostic characters and genetic markers. Syst. Parasitol. 45, 185–197. https://doi.org/10.1023/A:1006296316222 (2000).
Valentini, A. et al. Genetic relationships among Anisakis species (Nematoda: Anisakidae) inferred from mitochondrial cox2 sequences, and comparison with allozyme data. J. Parasitol. 92, 156–166 (2006).
Colón-Llavina, M. M. et al. Additional records of metazoan parasites from Caribbean marine mammals, including genetically identified anisakid nematodes. Parasitol Res. 105, 1239–1252 (2009).
Cavallero, S., Nadler, S. A., Paggi, L., Barros, N. B. & D’Amelio, S. Molecular characterization and phylogeny of anisakid nematodes from cetaceans from southeastern Atlantic coasts of USA, Gulf of Mexico, and Caribbean Sea. Parasitol. Res. 108, 781–792 (2011).
Kijewska, A., Dzido, J., Shukhgalter, O. & Rokicki, J. Anisakid parasites of fishes caught on the African shelf. J. Parasitol. 95, 639–645 (2009).
Quiazon, K. M. A., Santos, M. D. & Yoshinaga, T. Anisakis species (nematoda: Anisakidae) of dwarf sperm whale kogia sima (Owen, 1866) stranded off the pacific coast of southern Philippine archipelago. Vet. Parasitol. https://doi.org/10.1016/J.VETPAR.2013.05.019 (2013).
Zhang, L., Du, X., An, R., Li, L. & Gasser, R. B. Identification and genetic characterization of Anisakis larvae from marine fishes in the South China Sea using an electrophoretic-guided approach. Electrophoresis 34, 888–894 (2013).
Luo, H.-Y., Chen, H.-Y., Chen, H.-G. & Shih, H.-H. Scavenging hagfish as a transport host of anisakid nematodes. Vet. Parasitol. 218, 15–21. https://doi.org/10.1016/j.vetpar.2016.01.005 (2016).
Kuhn, T., Hailer, F., Palm, H. W. & Klimpel, S. Global assessment of molecularly identified Anisakis dujardin, 1845 (nematoda: Anisakidae) in their teleost intermediate hosts. Folia Parasitol. (Praha). 60, 123–134. https://doi.org/10.14411/fp.2013.013 (2013).
Grainger, J. N. R. The Identity of the larval nematodes found in the body muscles of the cod (Gadus callarias L.). Parasitology 49, 121–131 (1959).
Costa, G., Chada, T., Melo-Moreira, E., Cavallero, S. & D’Amelio, S. Endohelminth parasites of the leafscale gulper shark, Centrophorus squamosus (Bonnaterre, 1788) (Squaliformes: Centrophoridae) off Madeira archipelago. Acta Parasitol. 59, 316–322. https://doi.org/10.2478/s11686-014-0247-x (2014).
Hermida, M. et al. Infection levels and diversity of anisakid nematodes in blackspot seabream, Pagellus bogaraveo, from Portuguese waters. Parasitol. Res. 110, 1919–1928 (2012).
Sequeira, V. et al. Macroparasites as biological tags for stock identification of the bluemouth, Helicolenus dactylopterus (Delaroche, 1809) in Portuguese waters. Fish Res. 106, 321–328. https://doi.org/10.1016/j.fishres.2010.08.014 (2010).
Shamsi, S., Spröhnle-Barrera, C. & Shafaet, H. M. Occurrence of Anisakis spp. (Nematoda: Anisakidae) in a pygmy sperm whale Kogia breviceps (Cetacea: Kogiidae) in Australian waters. Dis. Aquat. Organ. 134, 65–74. https://doi.org/10.3354/dao03360 (2019).
Mcalpine, D. F., Murison, L. D. & Hoberg, E. P. New records for the pygmy sperm whale, Kogia breviceps (physeteridae) from Atlantic Canada with notes on diet and parasites. Mar. Mammal. Sci. 13, 701–704. https://doi.org/10.1111/j.1748-7692.1997.tb00093.x (1997).
Gunter, G. & Overstreet, R. Cetacean notes. I. Sei and rorqual whales on the Mississippi coast, a correction. II. A dwarf sperm whale in Mississippi sound and its helminth parasites. Gulf Res. Rep. 4, 479–481 (1974).
Mignucci-Giannoni, A. A., Hoberg, E. P., Siegel-Causey, D. & Williams, E. H. Metazoan parasites and other symbionts of cetaceans in the Caribbean. J. Parasitol. 84, 939–946 (1998).
Vidal, O., Findley, L. T., Turk, P. J. & Boyer, R. E. Recent records of pygmy sperm whales in the Gulf of California. Mexico. Mar. Mammal. Sci. 3, 354–356. https://doi.org/10.1111/J.1748-7692.1987.TB00323.X (1987).
Dollfus, R. P. Helminthofaune de Kogia breviceps (Blainxille, 1938) cetace odontocete. Recoltes du Dr R. Duguy. Ann. Sci. Natl. Charente-Maritime 4, 3–6 (1966).
MCAlpine, D.F., (2018) Pygmy and dwarf sperm whales. In: Encyclopedia of Marine Mammals. Elsevier p. 786–8.
Fernández, R., Santos, M. B., Carrillo, M., Tejedor, M. & Pierce, G. J. Stomach contents of cetaceans stranded in the canary Islands 1996–2006. J. Mar. Biol. Assoc. United Kingdom. 89, 873–883 (2009).
Berrow, S., López Suárez, P., Jann, B., Ryan, C., Varela, J., Hazevoet, C.J., (2015) Recent and noteworthy records of Cetacea from the Cape Verde Islands. www.scvz.org. Accessed 1 Mar 2021.
Mattiucci, S., Nascetti, G., (2008) Chapter 2 advances and trends in the molecular systematics of anisakid nematodes, with implications for their evolutionary ecology and host-parasite co-evolutionary processes. Adv. Parasitol. 66 47 148
Measures, L.N., (2014) Anisakiosis and pseudoterranovosis. Reston, Virginia; https://doi.org/10.3133/cir1393
McClelland, G. The trouble with sealworms (Pseudoterranova decipiens species complex, nematoda): A review. Parasitology 2002(124 Suppl), S183-203 (2009).
Alt, K. G., Cunze, S., Kochmann, J. & Klimpel, S. Parasites of three closely related Antarctic fish species (teleostei: Nototheniinae) from Elephant Island. Acta Parasitol. https://doi.org/10.1007/s11686-021-00455-8 (2021).
McClelland, G. Phocanema decipiens (Nematoda: Anisakinae): Experimental infections in marine copepods. Can. J. Zool. 60, 502–509. https://doi.org/10.1139/z82-075 (1982).
Marcogliese, D. J. Review of experimental and natural invertebrate hosts of sealworm (Pseudoterranova decipiens) and its distribution and abundance in macroinvertebrates in eastern Canada. NAMMCO Sci. Publ. 3, 27–37 (2001).
West, K. L. et al. Diet of pygmy sperm whales (Kogia breviceps) in the Hawaiian Archipelago. Mar. Mammal. Sci. 25, 931–943. https://doi.org/10.1111/j.1748-7692.2009.00295.x (2009).
Kleinertz, S., Damriyasa, I. M., Hagen, W., Theisen, S. & Palm, H. W. An environmental assessment of the parasite fauna of the reef-associated grouper Epinephelus areolatus from Indonesian waters. J. Helminthol. 88, 50–63 (2014).
Nadler, S. A. et al. Molecular phylogenetics and diagnosis of Anisakis, Pseudoterranova, and Contracaecum from northern pacific marine mammals. J. Parasitol. 91, 1413–1429 (2005).
Weitzel, T. et al. Human infections with Pseudoterranova cattani nematodes. Chile. Emerg. Infect. Dis. 21, 1874–1875 (2015).
Arizono, N., Miura, T., Yamada, M., Tegoshi, T. & Onishi, K. Human infection with Pseudoterranova azarasi roundworm. Emerg. Infect. Dis. 17, 555–556. https://doi.org/10.3201/eid1703.101350 (2011).
Kleinertz, S. et al. Gastrointestinal parasites of free-living Indo-Pacific bottlenose dolphins (Tursiops aduncus) in the Northern Red Sea. Egypt. Parasitol Res. 113, 1405–1415. https://doi.org/10.1007/s00436-014-3781-4 (2014).
Aco Alburqueque, R., Palomba, M., Santoro, M. & Mattiucci, S. Molecular identification of zoonotic parasites of the genus Anisakis (Nematoda: Anisakidae) from fish of the southeastern Pacific Ocean (off Peru coast). Pathogens. 9, 910. https://doi.org/10.3390/pathogens9110910 (2020).
Di Azevedo, M. I. N., Carvalho, V. L. & Iñiguez, A. M. Integrative taxonomy of anisakid nematodes in stranded cetaceans from Brazilian waters: An update on parasite’s hosts and geographical records. Parasitol. Res. 116, 3105–3116. https://doi.org/10.1007/s00436-017-5622-8 (2017).
Quiazon, K. M. A., Santos, M. D., Blatchley, D. D., Aguila, R. D. & Yoshinaga, T. Molecular and morphological identifications of Anisakis dujardin, 1845 (Nematoda: Anisakidae) from a rare deraniyagala’s beaked whale (Mesoplodon hotaula deraniyagala, 1963) and blainville’s beaked whale (Mesoplodon densirostris blainville, 1817) stranded. Philipp. J. Sci. 150, 823–835 (2021).
Bao, M. et al. Air-dried stockfish of Northeast Arctic cod do not carry viable anisakid nematodes. Food Cont. 116, 107322. https://doi.org/10.1016/j.foodcont.2020.107322 (2020).
Liu, G. H. et al. Mitochondrial phylogenomics yields strongly supported hypotheses for ascaridomorph nematodes. Sci. Rep. https://doi.org/10.1038/srep39248 (2016).
Hrabar, J. et al. Phylogeny and pathology of anisakids parasitizing stranded California sea lions (Zalophus californianus) in Southern California. Front Mar. Sci. https://doi.org/10.3389/fmars.2021.636626 (2021).
Acknowledgements
We thank Natalia Drivenes for the excellent technical support and help with the molecular analyses. The authors thank the captain and the crew on board the R.V. “Kronprins Haakon”, and the colleagues who participated in the cruise. We acknowledge the projects HARMES (Research Council of Norway project number 280546) and MEESO (EU H2020 research and innovation programme, Grant Agreement No 817669) for supporting the sampling collection.
Funding
The study was internally financed by the IMR.
Author information
Authors and Affiliations
Contributions
M.B., A.L., P.C. and E.K. were involved in the conception and design of the study. M.B. and E.K. supervised K.M.O., who carried out the parasite sampling, morphological and molecular identification, and took parasite images. M.B. and E.K. analysed and interpreted the data and wrote the original manuscript. L.G. and J.E.S. helped supervising the molecular analyses. E.G.S. participated in sample collection and identification of the fish species. All authors reviewed and contributed to the manuscript and approved its final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bao, M., Olsen, K.M., Levsen, A. et al. Characterization of Pseudoterranova ceticola (Nematoda: Anisakidae) larvae from meso/bathypelagic fishes off Macaronesia (NW Africa waters). Sci Rep 12, 17695 (2022). https://doi.org/10.1038/s41598-022-22542-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22542-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.