Abstract
Parasite biodiversity in cetaceans represents a neglected component of the marine ecosystem. This study aimed to investigate the distribution and genetic diversity of anisakid nematodes of the genus Anisakis sampled in cetaceans from the Northeast Atlantic Ocean and the Mediterranean Sea. A total of 478 adults and pre-adults of Anisakis spp. was identified by a multilocus genetic approach (mtDNA cox2, EF1 α − 1 nDNA and nas 10 nDNA gene loci) from 11 cetacean species. A clear pattern of host preference was observed for Anisakis spp. at cetacean family level: A. simplex (s.s.) and A. pegreffii infected mainly delphinids; A. physeteris and A. brevispiculata were present only in physeterids, and A. ziphidarum occurred in ziphiids. The role of cetacean host populations from different waters in shaping the population genetic structure of A. simplex (s.s.), A. pegreffii and A. physeteris was investigated for the first time. Significant genetic sub-structuring was found in A. simplex (s.s.) populations of the Norwegian Sea and the North Sea compared to those of the Iberian Atlantic, as well as in A. pegreffii populations of the Adriatic and the Tyrrhenian Seas compared to those of the Iberian Atlantic waters. Substantial genetic homogeneity was detected in the Mediterranean Sea population of A. physeteris. This study highlights a strong preference by some Anisakis spp. for certain cetacean species or families. Information about anisakid biodiversity in their cetacean definitive hosts, which are apex predators of marine ecosystems, acquires particular importance for conservation measures in the context of global climate change phenomena.
Similar content being viewed by others
Introduction
Anisakid nematodes of the genus Anisakis Dujardin, 1845 are marine parasites with a worldwide distribution. Their life cycle is indirect, involving several hosts at different trophic levels of marine food webs. The parasite’s adult stage lives in marine mammals, mainly cetaceans, while planktonic or semi-planktonic crustaceans act as first intermediate hosts of the parasite, and fish and squid represent intermediate/paratenic hosts1,2,3. Specifically, Anisakis spp. parasitize the digestive tracts of cetaceans, being commonly found in the stomachs of both toothed and baleen whales1,3,4,5,6,7. The definitive hosts acquire Anisakis through ingestion of infected crustaceans, fish or squid. Once in the stomach of cetacean hosts, the third larval stages (L3) of Anisakis spp. moult into the fourth stage larvae (L4), that further develop into sexually mature adults, a process taking from 40 to 60 days8,9,10. The adult stages of Anisakis “swim” in the ingesta, probably feeding on them9. In many cases they attach themselves to the stomach mucosa, forming clusters and sometimes generating gastric granulomatous ulcers11,12,13,14,15.
The application of genetic/molecular methodologies to Anisakis morphospecies has advanced our understanding of their systematics, taxonomy, and phylogeny, allowing the detection and description of sibling species and the discovery of new species2,3,16,17,18, and generally furthering our knowledge of their ecology, geographical distribution and host preferences1,3,19,20,21,22. The existence of nine nominal species of Anisakis as distinct phylogenetic units has been demonstrated by various phylogenetic analyses, using both nuclear and mitochondrial genes2,3,19,23,24. PCR–RFLP of the ITS rDNA region24 and the sequencing of the mitochondrial mtDNA cox2 gene25 are used to identify all the species of Anisakis, at both larval and adult stages. Several nuclear gene loci have been recently validated on specimens of the A. simplex s.l. complex (i.e. A. pegreffii, A. simplex (s.s.) and A. berlandi): sequence analysis of the EF1 α − 1 nDNA region revealed two diagnostic nuclear sites (SNPs) at which the nucleotides differed between A. simplex (s.s.) and A. pegreffii26; the metallopeptidase 10 (nas10 nDNA) locus exhibits diagnostic SNPs, at which the nucleotides differ between the three species, and for which ARMS-PCR assays were developed27. Panels of DNA microsatellite (SSR) loci have been developed in recent years17,28, which include some 100% diagnostic loci with fixed alternative alleles which differ between the three species17,29,30.
Studies on the epidemiology of Anisakis spp. in their intermediate/paratenic hosts (i.e. fish and squid) from Northeast (NE) Atlantic Ocean and Mediterranean Sea are abundant, and this field is regularly updated3,31,32,33,34,35. However, there is scant epidemiological data on Anisakis spp. in their definitive hosts (i.e., cetaceans) and consequently knowledge is lacking on their biodiversity, distribution, and ecology at the pre-adult and adult stages. To date, investigations on parasites of cetaceans in the NE Atlantic and Mediterranean Sea have relied on samples from stranded individuals3,12,14,15,36,37,38,39 or, more rarely, on samples obtained during whaling activities8.
Knowledge of the biodiversity (at both species and genus level) of Anisakis in different cetacean hosts has important implications for the understanding of the parasite/host ecology. The aim of the present work was to apply a multilocus genetic approach to the study of anisakid endoparasites of cetaceans from European seas, to: (i) assess the biodiversity of Anisakis species at their adult stage in various cetacean hosts, from different areas of the NE Atlantic Ocean and the Mediterranean Sea; (ii) provide insights into the distribution of Anisakis species in cetacean populations; (iii) analyse the host-parasite association between cetacean hosts and their Anisakis parasites; (iv) investigate the population genetic structure of Anisakis spp. from different hosts and geographical ranges.
Results
Parasite samples
A total of N = 478 anisakid nematodes was obtained from stranded cetaceans and, according to their morphological characteristics, they were all assigned to the Anisakis genus. Only adult (showing developed reproductive structures, such as caudal papillae and spicules in male worms and ovaries in females) and pre-adult (presence of labia, absence of boring tooth) specimens were found. It was not possible to count or estimate the total burden of worms during most of the necropsies of cetaceans, only to obtain a subsample of parasites. Hence, we could not perform a comprehensive epidemiological analysis of the parasites for each host species.
Genetic identification of Anisakis
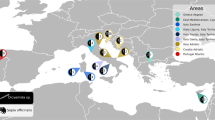
According to the BLAST analysis performed on the 478 mtDNA cox2 sequences (580 bp) obtained, five species were detected: A. simplex (s.s.), A. pegreffii, A. physeteris, A. brevispiculata and A. ziphidarum. The numbers of Anisakis specimens genetically identified in the definitive hosts from different sampling area are reported in Table 1. The relative proportions of Anisakis spp. in cetaceans in different geographical areas of the NE Atlantic Ocean and the Mediterranean Sea, and in their different definitive hosts families, are reported in Figs. 1 and 2, respectively.
Relative proportions of Anisakis spp. in cetaceans according to different geographical areas of the NE Atlantic Ocean and the Mediterranean Sea. The map was obtained from Wikimedia Commons, licensed-free (https://commons.wikimedia.org/wiki/File:BlankMap-Europe-v4.png), Roke was the unofficial uploader of the derivitive map CC BY-SA 3.0.
Relative proportions of Anisakis spp. according to definitive hosts belonging to species of families Balenopteridae, Delphinidae, Phocoenidae, Physeteridae and Ziphiidae. Host codes are reported by alphabetical order: Ba: Balenoptera acutorostrata, Dd: Delphinus delphis, Gm: Globicephala melas, La: Lagenorhynchus albirostris, Tt: Tursiops truncatus, Sc: Stenella coeruleoalba, Pp: Phocoena phocoena, Pm: Physeter macrocephalus, Ks: Kogia sima, Kb: Kogia breviceps, Zc: Ziphius cavirostris.
The sympatric (in some cases even syntopic, such as in two specimens of common dolphins Delphinus delphis, one striped dolphin Stenella coeruleoalba, one bottlenose dolphin Tursiops truncatus and three harbour porpoises Phocoena phocoena) occurrence of 134 adult specimens of A. pegreffii and A. simplex (s.s.) was recorded from dolphin species stranded along the Iberian Atlantic coast (Table 1). These specimens were also identified using nuclear diagnostic loci, i.e. the EF1 α − 1 nDNA and nas10 nDNA loci. Among them, 134 specimens showed 100% concordance with the mtDNA cox2 data set for the identification of the two parental taxa, i.e. A. simplex (s.s.) (N = 107) and A. pegreffii (N = 27). Indeed, at the EF α − 1 nDNA locus, 107 individuals were found to be homozygous C/C and T/T, respectively, at the fixed alternative nucleotide positions 186 and 286, i.e. diagnostic for the parental species A. simplex (s.s.). A further 25 showed the homozygote genotypes T/T and C/C at the same SNPs, corresponding to the parental species A. pegreffii26 (Suppl. Fig. 1). Additionally, the ARMS-PCR analysis enabled us to genotype 107 individuals as belonging to A. simplex (s.s.), and 25 to the species A. pegreffii. The combined use of the nas 10 primers generated a specific PCR product of 296 bp in A. simplex (s.s.), amplifying the T-allele, and a specific PCR fragment of 117 bp in A. pegreffii, amplifying the C-allele. In contrast, two specimens obtained respectively from a harbour porpoise and a short-beaked common dolphin, which at the mtDNA cox2 locus corresponded to the species A. pegreffii, showed a heterozygote pattern at those diagnostic positions (SNPs)—thus falling between A. pegreffii and A. simplex (s.s.), in both the two nuclear loci EF1 α − 1 nDNA and nas10 nDNA (Suppl. Fig. 1). In particular, the sequence analysis of these two specimens revealed the occurrence of a double peak at these diagnostic positions (i.e., 186 and 286) at the EF1 α − 126. For the ARMS analysis, two specific PCR fragments of both 296 and 117 bp were generated in the species with a heterozygote pattern, amplifying both the C and T allele. This latter finding showed 100% concordance with the direct sequence analysis of the gene locus nas 10 nDNA. These two specimens, showing a heterozygous pattern at all the diagnostic SNPs of the EF1 α − 1 and nas10 nDNA loci, seem to be the result of hybridization events between A. pegreffii and A. simplex (s.s.).
The DNA sequences of the presently identified Anisakis species were deposited in GenBank under the following accession numbers: mtDNA cox2: OM142467–OM142471 (A. pegreffii); OM142472–OM142476 (A. simplex (s.s.)); OM14247–OM142481 (A. physeteris). EF1 α − 1 nDNA: OM210033–OM210037 (A. pegreffii); OM210038 (A. simplex (s.s.)).
Phylogenetic analysis inferred from mtDNA cox2 gene locus
The Bayesian inference (BI) phylogenetic tree based on mtDNA cox2 sequences (Fig. 3) indicated that a total of 220 nematodes, including preadult and adult specimens recovered in minke whale (Balaenoptera acutorostrata) and six delphinid species (striped dolphin Stenella coeruleoalba, bottlenose dolphin Tursiops truncatus, harbour porpoises Phocoena phocoena, long-finned pilot whale Globicephala melas, white-beaked dolphin Lagenorhynchus albirostris, and common dolphin Delphinus delphis) and a few preadults found in physeterids (sperm whale Physeter macrocephalus and pygmy sperm whale Kogia breviceps), clustered together within a well-supported clade (100% probability value), together with a reference sequence of A. simplex (s.s.) (DQ116426) (Fig. 3). Similarly, a total of 96 Anisakis preadult and adult specimens grouped in a highly (100%) supported phylogenetic lineage represented by A. pegreffii, which also included a reference sequence (DQ116428) of the parasite species (Fig. 3). Additionally, 157 specimens collected in physeterids from the Mediterranean Sea and the Atlantic Ocean clustered in a well-supported distinct phylogenetic clade formed by the species A. physeteris (Fig. 3). Two adult Anisakis specimens collected from pygmy sperm whale (Kogia breviceps) in Atlantic waters clustered in a clade represented by A. brevispiculata (Fig. 3, Table 1). Two adult worms collected from Cuvier's beaked whale (Ziphius cavirostris) from the Aegean Sea were placed in a highly supported clade comprising sequences of A. ziphidarum previously deposited in GenBank (Fig. 3, Table 1).
Phylogenetic tree of cox2 mtDNA gene locus constructed using Bayesian Markov Chain Monte Carlo (MCMC) analysis in BEAST v1.10.4109. GTR + I + G substitution model, strict molecular clock103 and Yule speciation process were used as the tree priors, Bayesian inference (BI) on mtDNA cox2 gene sequences of Anisakis spp. obtained from cetaceans. Toxocara canis and Ascaris suum were used as an outgroup.
Genetic diversity inferred from mtDNA cox2 gene locus
The genetic diversity analysis was performed at intraspecific level in A. simplex (s.s.), A. pegreffii and A. physeteris. It was not possible to give estimates of genetic diversity of A. ziphidarum and A. brevispiculata because of the low number of worms identified for those species.
The hierarchical gene diversity analysis by AMOVA (Suppl. Table 1) indicated that in A. simplex (s.s.) 92.77% of the genetic variation was expressed within populations and only 2.52% was variation among populations of this parasite species collected from cetacean hosts of different geographic areas of the NE Atlantic Ocean (Suppl. Table 1). Similar results were obtained for A. pegreffii, with 97.48% of variation occurring within populations of the parasite species collected in cetaceans from different areas (Suppl. Table 1). In the case of A. physeteris 94.62% of variation occurred within populations (Suppl. Table 1).
Estimates of the genetic differentiation (Fst) between populations of the presently identified Anisakis species, as inferred from the fixation index, are shown in Table 2. At the intraspecific level, a significant genetic differentiation (Fst) was found between A. simplex (s.s.) metapopulations from the Norwegian Sea compared to those from Iberian Atlantic coast (Fst ≈ 0.12, p < 0.00001), as well as between populations of the same species from the Scottish and Iberian Atlantic coast (Fst ≈ 0.07, p < 0.00001). However, A. simplex (s.s.) individuals collected from the Norwegian Sea had a lower pairwise Fst when compared to its conspecifics in Scottish waters (Fst ≈ 0.02, p = 0.03) (Table 2). Comparing host species, significant Fst values were found between A. simplex (s.s.) metapopulations from long-finned pilot whales (Faroes Islands and Iberian Atlantic) and those collected from the minke whale from Norwegian Sea (Fst ≈ 0.10, p < 0.00001) and white-beaked dolphin from Scottish waters (Fst ≈ 0.07, p < 0.00001) (data not shown). However, a higher Fst pairwise value (Fst ≈ 0.15, p < 0.001) was recorded when comparing parasite metapopulations from the same cetacean species, i.e. pilot whale subpopulation from Faroes Island and Iberian Atlantic coast. Similarly, a significant genetic differentiation was found when comparing the metapopulations of A. simplex (s.s.) from striped dolphins of Iberian Atlantic coast and Scottish waters (Fst ≈ 0.09, p < 0.00001).
Intraspecific genetic variation between metapopulations of A. pegreffii from the Adriatic and the Tyrrhenian Seas was not significant (Fst ≈ 0.015, p = 0.10) (Table 2). In contrast, significant genetic sub-structuring was observed when comparing the Mediterranean (Adriatic and Tyrrhenian) populations of A. pegreffii with those from the Iberian Atlantic coast (Fst ≈ 0.07 (p = 0.0001) and Fst ≈ 0.04 (p = 0.001), respectively (Table 2)). Due to the low number of A. pegreffii found in the different definitive hosts examined, except for striped dolphins, it was not possible to compare the Fst value at intraspecific level by host species.
Due to the low number of specimens of A. physeteris obtained from the NE Atlantic Ocean, Fst at the intraspecific level was calculated only for the comparison of specimens collected from stranded sperm whales along the coast of the Adriatic Sea (of Ionian Sea origin31) and those carried by the dwarf sperm whale (Kogia sima) from the Tyrrhenian Sea, with the finding of a generally low and slightly significant level of differentiation (Fst ≈ 0.012, p = 0.03) (Table 2).
At the interspecific level, significant values of genetic differentiation were observed between A. simplex (s.s.) and A. pegreffii (Fst ≈ 0.836), with the highest values being those found between A. physeteris and A. simplex (s.s.) (Fst ≈ 0.945) and between A. physeteris and A. pegreffii (Fst ≈ 0.940).
The haplotype parsimony network (TCS) inferred from mtDNA cox2 haplotypes of A. simplex (s.s.) (Fig. 4) shows that the presence of two major haplotypes, i.e. Hap25 and Hap48. Hap25 was shared by several A. simplex (s.s.) individuals collected in the cetacean hosts from the Norwegian Sea and Scottish coast of the NE Atlantic Ocean, whereas Hap48 was shared mostly by the A. simplex (s.s.) metapopulations collected in the Iberian and Scottish Atlantic (Fig. 4). The TCS analysis revealed the existence of several unique haplotypes in the Norwegian metapopulation of A. simplex (s.s.) and in Iberian Atlantic waters. Moreover, TCS also shows concordance between the haplotype clustering and geographic origin of the endoparasite samples (Fig. 4).
TCS analysis of mtDNA cox2 haplotypes of A. pegreffii resulted in a star-like tree (Fig. 5), with Hap3 being the most frequent haplotype, shared by all the metapopulations of the parasite (Fig. 5). A high frequency of certain unique haplotypes in the parasite specimens from the definitive hosts stranded on the Spanish Atlantic coast and from the Adriatic Sea was observed in A. pegreffii (Fig. 5).
Finally, results from the TCS analysis of A. physeteris haplotypes did not show grouping by geographic regions (Fig. 6), but instead revealed a star-shaped pattern, with Hap8 (likely the ancestral haplotype) being the most frequent haplotype, shared by specimens from the Ionian Sea and the Tyrrhenian Sea. The analysis also showed the existence of many haplotypes in several areas of the Mediterranean Sea, differentiated from each other by a few substitutions (Fig. 6). The haplotype diversity (h) of A. simplex (s.s.), estimated by the analysis of mtDNA cox2 sequences, was high, ranging between 0.97 and 0.98 in parasite metapopulations from Iberian Atlantic waters and the Norwegian Sea, respectively (Table 3). Slightly lower values were found in A. pegreffii (on average, h ≈ 0.95) and in A. physeteris (on average, h ≈ 0.95) (Table 3). The average value of nucleotide diversity within species was π ≈ 0.007, π ≈ 0.005 and π ≈ 0.006 in A. simplex (s.s.), A. pegreffii and, A. physeteris, respectively (Table 3).
Discussion
Identification of Anisakis spp. in cetaceans
Knowledge of the occurrence and distribution of adult stage worms of the genus Anisakis in their definitive hosts is mostly based on opportunistic samples, obtained during necropsy investigations of cetaceans which were stranded, bycaught in fishing gear or taken by whaling6,8,40. Use of stranding records is a valid and sustainable approach to study the diversity of local marine mammal species41,42,43,44,45, and to investigate the parasite fauna hosted by these animals. Stranding events can occur for several reasons, including natural causes such as illness, environmental factors, and anthropogenic causes including bycatch, vessel strikes, plastic ingestion, or acoustic trauma46,47,48,49,50,51,52,53,54. Despite these advantages, parasitological studies on stranded cetaceans are affected by the decomposition state of the animals. It is also possible that animals in poor nutritional condition or moribund due to disease or trauma are unlikely to have been successfully foraging before death, which may affect the parasite burden demonstrated at necropsy.
In this study, five Anisakis species, namely A. simplex (s.s.), A. pegreffii, A. physeteris, A. brevispiculata and A. ziphidarum, were detected from 11 species of cetacean stranded at various locations along the coasts of the NE Atlantic Ocean and Mediterranean Sea (Fig. 7, Table 4). In addition, a multilocus genotyping approach based on nuclear markers, applied to specimens of the sibling species A. simplex (s.s.) and A. pegreffii obtained from hosts occurring in a sympatric area (Iberian Atlantic waters), allowed the identification of two heterozygote genotypes, i.e. two F1 hybrids, showing a heterozygous pattern at all of the diagnostic nucleotide positions observed at the EF1 α − 1 nDNA and nas 10 nDNA loci (Suppl. Fig. 1). This finding suggests that these heterozygote genotypes most likely originated from a recent hybridization event between sympatric specimens of A. simplex (s.s.) and A. pegreffii. There have been reports of A. simplex (s.s.) and A. pegreffii L3 heterozygote genotypes in the same sympatric area from several fish species, including species which are likely to be eaten by the studied cetaceans26,29,55,56,57,58,59,60,61. Thus, hybridization events between species of the A. simplex (s. l.) complex are probably a common phenomenon in sympatric areas, as also recently reported from southern oceans based on a multinuclear genotyping approach29.
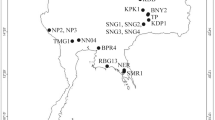
Sampling localities of Anisakis spp. in the Mediterranean Sea and NE Atlantic Ocean of the occasional cetacean strandings, which resulted in 34 specimens belonging to 11 species. The map was obtained from Wikimedia Commons, licensed-free (https://commons.wikimedia.org/wiki/File:BlankMap-Europe-v4.png), Roke was the unofficial uploader of the derivitive map CC BY-SA 3.0.
The phylogenetic tree topology, based on the mtDNA cox2 sequences obtained (Fig. 3), differentiated Anisakis spp. into four well-defined clades, in accordance with previous studies3,23. The first clade comprised species of the A. simplex complex, i.e. A. simplex (s.s.), A. pegreffii (and A. berlandi, not found in the current study). The second clade included A. ziphidarum and A. nascettii (the latter not found in the current study), while the third clade comprised A. physeteris, A. brevispiculata, and A. paggiae (the latter not found in the current study).
Host preference by Anisakis spp.
The well-known existence of a host specificity pattern in A. simplex (s.s.) and A. pegreffii for dolphins and baleen whales has been further supported by the present findings. Indeed, the three sibling species of the A. simplex (s. l.) complex are primarily parasites of the cetacean families Delphinidae, Monodontidae, Phocoenidae and Balaenopteridae1,2,6,8,19,23,38,62, parasitizing these hosts in different geographical areas, depending on the range of distribution of both host and parasite species3,7,30.
In the current study, adult nematodes of A. simplex (s.s.) and A. pegreffii were found in all the species of delphinids that were examined and in a minke whale specimen (Table 1, Figs. 1 and 2). Adult parasites of these two species can often be found in syntopy in the same definitive hosts in areas where the geographical range of the two parasite species overlaps26,60, as observed in bottlenose dolphins, striped dolphins, common dolphins and harbour porpoises inhabiting the waters off the Iberian coast (Table 1, Figs. 1 and 2). A sympatric and syntopic distribution of these two species was previously observed in other delphinids and whales sampled along the Japanese coasts, such as bottlenose dolphin, minke whale, and sei whale (B. borealis)6. Similarly, a syntopic occurrence of A. pegreffii and A. berlandi was reported in long-finned pilot whales from New Zealand30, as well as in a killer whale (Orcinus orca) from Argentinian waters7. Another relevant observation concerning the preference of these parasite species for certain definitive hosts is the different size reached by mature specimens of A. simplex (s.l.) in mysticetes, killer whales and smaller dolphins6,8. Gomes et al.6, and Ugland et al.8, reported that A. simplex (s.s.) and A. pegreffii specimens seem to reach bigger sizes in mysticetes (sei whales and minke whales) than in Delphinidae (bottlenose dolphins, long finned pilot whales and striped dolphins). Bigger whales probably offer a more beneficial microhabitat to these parasite species compared to smaller delphinids, enhancing their fitness potential. However, the coevolutionary implications related to these observations have not yet been clarified.
This investigation further confirms that cetaceans of the families Physeteridae (i.e. sperm whale) and Kogiidae (i.e. dwarf sperm whale, K. sima, and pygmy sperm whale, K. breviceps) are the main definitive hosts for the species included in clade 3 (Figs. 1, 2, 3), comprising A. physeteris, A. brevispiculata and A. paggiae. However, a very rare infection of (preadult) A. simplex (s.s.) and A. pegreffii was recorded in physeterids in the present study. Santoro et al.63 reported the presence of a few immature specimens of A. pegreffii in syntopy with fully developed adult specimens of A. physeteris located in the stomach of the same specimen of dwarf sperm whale examined in the present study. A rare case of infection with A. simplex (s.s.) in a pygmy sperm whale from the Caribbean Sea was also previously observed19, but the developmental stage of the worms was not reported. The rare cases of co-infection with both A. simplex (s.s.) and A. pegreffii in physeterid species, mostly at the preadult stage, strengthen the hypothesis that infections with these two parasite species in physeterids are quite uncommon and probably accidental. In this study, A. physeteris was found to be the most common anisakid in the sperm whale, representing 97% of the identified specimens, and 82% of the anisakids recovered from dwarf sperm whale. These proportions are similar to those previously reported in other studies39,64.
The host preference of A. physeteris for odontocetes belonging to the family Physeteridae, specifically the sperm whale, could be related to the ecology and the peculiar life cycle of this Anisakis species. A. physeteris has been found sporadically in fish species, with low infection rates3,34,35 probably because it relies on deep-sea squid species as transport/paratenic hosts65. Large, deep-dwelling cephalopods may act as reservoir hosts for this anisakid species, and may thus be crucial to complete its life cycle and maintain its distribution. In fact, the whole life cycle of this parasite species is probably strictly connected to deep sea ecosystems. Considering the feeding behaviour of the sperm whales, the role of squid of the family Histiotheutidae could be crucial in the life cycle of A. physeteris, as recently suggested by the finding of A. physeteris larvae in Histioteuthis bonnellii in the depths of the Central Mediterranean Sea (Tyrrhenian Sea)65. Thus, deep-dwelling squid species, which are common prey of physeterid whales39,43,66,67,68,69, could be a suitable paratenic/transport host for this Anisakis species. The stomachs of three out of the six sperm whales stranded along the Italian coast of the Adriatic Sea in 2017 showed a mean abundance of 706 specimens of A. physeteris per whale39. In the same stomachs, along with this burden of preadult and adult specimens of A. physeteris, over 4000 cephalopods beaks were counted, for each whale. Of these beaks, 95% were classified as belonging to histioteuthids (71% H. bonnellii and 24% H. reversa), while the rest were Ancistrocheirus lesueurii (Ancistrocheiridae) and Octopoteuthis sicula (Octopoteuthidae)39. The co-occurrence of pre-adults and adults of A. physeteris along with large numbers of squid beaks in the above-mentioned stomachs indicates a possible link between these deep-sea squid species, probably carrying A. physeteris parasites in high numbers. A similar finding was reported from a sperm whale stranded along the Southeastern coast of Italy, which was parasitized by several hundred adult A. physeteris70 and had the stomach full of beaks of the squids H. bonnellii and Ancistroteuthis lenchisteini (Mattiucci and Cipriani pers. obs.).
It could be hypothesized that A. pegreffii and/or A. simplex (s.s.) were ingested along with A. physeteris. The latter, being adapted to the specific stomach-microhabitat of physeterid hosts, could be more competitive. Similar dynamics have been observed in several other sibling species of anisakid nematodes71 as well as in other helminth parasites72.
Finally, two A. ziphidarum adult specimens, identified from a Cuvier´s beaked whale (Ziphius cavirostris) stranded on the Italian coast of the Ionian Sea, clustered in Clade 2 (Fig. 3), along with the closely related species A. nascettii. To date, A ziphidarum is the only anisakid species found in Cuvier's beaked whale from the Mediterranean Sea73 while A. nascettii (not found in the current study) was mainly reported from beaked whales of the genus Mesoplodon4,74. The feeding habits of beaked whales suggest that the life cycles of A. ziphidarum and A. nascettii may mostly involve deep-sea intermediate hosts such as deep-sea squids and mesopelagic fish. Indeed, ziphiids commonly prey on squid belonging to the families Onychoteuthidae and Histiotheuthidae, rather than fish75,76,77.
The association observed between Anisakis spp. and several cetacean taxa may reflect a co-evolutionary history between these endoparasites and their hosts over the millennia, driven by common trophic adaptations. In particular, the distinct clades formed by the species of Anisakis have been suggested to “mirror” the clades so far reported in the phylogenetic analysis of their main definitive hosts2,21.
Drivers of the distribution of Anisakis spp. in cetacean species of European waters
Among the drivers that shape the geographical range of the various Anisakis species, particular consideration should be given to a dispersal mechanism involving both their definitive (cetaceans) and intermediate/paratenic (fish and squid) hosts.
The infection pattern of the adult Anisakis specimens recorded in their final hosts (Fig. 1) was largely in accordance with their documented distribution in intermediate/paratenic hosts from the Atlantic Ocean and the Mediterranean Sea3,35.
Generally, in the NE Atlantic Ocean, A. simplex (s.s.) and A. pegreffii are distributed along a latitudinal gradient1,3. In this study, we confirmed that A. pegreffii is the most widespread parasite species occurring in cetaceans from the Mediterranean Sea. In the NE Atlantic Ocean, the prevalence of A. pegreffii progressively declines northwards along the NorthWest Spanish coast (Fig. 1) and is absent from hosts from northern waters. An opposite geographic trend was found in A. simplex (s.s.), which showed a south-to-north increase in abundance when in sympatry with A. pegreffii in cetaceans stranded on the coast of Spanish Galicia, and was the only Anisakis species present in the cetaceans from the NE Atlantic Ocean (Fig. 1). A. simplex (s.s.) is the only Anisakis species so far reported from cetaceans in the northern NE Atlantic Ocean. This is confirmed by a recent study on Anisakis spp. in stranded harbour porpoises from the Norwegian Sea whose were identified molecularly as individuals of A. simplex (s.s.)15. In the same host, a study on specimens stranded on the North Sea, Baltic Sea and North Atlantic coasts, detected the presence of A. simplex (s.s.), as well as the presence of accidental parasites, e.g. Pseudoterranova decipiens (s.s.) (usually associated with pinnipeds as definitive hosts) and Hysterothylacium aduncum (a parasite maturing in teleost fish)78. Anisakis sp. was previously detected in 24 minke whales from the NE Atlantic Ocean, from which a large number of nematodes was recovered during whaling operations in the Barents Sea8. The worms were not molecularly identified at species level but presumably consisted of A. simplex (s.s.)23. A high prevalence of infection with adults of Anisakis in long-finned pilot whales was also reported from Faroe Islands8,79, even if identification was not provided. Mattiucci et al.23 identified several adults of A. simplex (s.s.) obtained from this same host in the Norwegian Sea.
In the study of genetic architecture drivers of Anisakis spp., the high dispersal capacity of some definitive and intermediate/paratenic hosts species (mainly pelagic fish) has a crucial role in maintaining gene flow between parasite populations. Indeed, some cetacean hosts are capable of remarkable migrations to feed or mate, carrying their parasites with them. However, it should be considered that larval Anisakis specimens in highly migratory fish species may have even a wider dispersal potential, since the larvae may remain latent for years9,80. Larval Anisakis spp. can live for years in their transport hosts9,80, while they probably need around 40–60 days to mature in their definitive hosts8,81, Mattiucci pers. obs. from in vitro culture experiments]. Highly migratory hosts, particularly in periods of intense migrations (mating or feeding), can spread their parasites over broad spatial scales, dispersing specimens even out of the geographical range in which they could find favourable ecological conditions for completing their life cycle. These migrating fish could be predated by suitable definitive hosts in areas far away from the original infection, where abiotic conditions, depending on their suitability for completion of the parasite life cycle, would represent a limiting factor for parasite dispersal.
For instance, A. simplex (s.s.) specimens have been rarely detected in the Mediterranean Sea. The few reports available34,35, of larval stages found in fish, refer mainly to the Alboran Sea, which is considered ecologically more similar to the Atlantic waters than to the rest of the Mediterranean Sea. However, three adult specimens of A. simplex (s.s.) were identified by sequences analyses of the ITS region of rDNA and mtDNA cox2 from bottlenose dolphin and striped dolphin, as well as two larval specimens from bluefin tuna (Thunnus thynnus) in the Adriatic Sea28,38,82,83. The presence of A. simplex (s.s.) outside the usual limits of its geographical range may be related to the wide migratory capability of the intermediate and definitive host species in which the parasite has been recorded. Hence, these cetaceans and pelagic fish species could have acquired the infection in areas within the usual range of A. simplex (s.s.) and these findings may not be indicative of the “true” habitat where these species could complete their life cycle.
A similar hypothesis could explain the rare findings of larval A. pegreffii in fish from the northern waters of the NE Atlantic Ocean, although they have not so far been found as adults in cetacean hosts from the same waters. Larvae of A. pegreffii were detected in the highly migratory Atlantic mackerel (Scomber scombrus) caught in the Norwegian Sea84, also in Atlantic cod (Gadus morhua) from the North Sea85, the latter known for being an active predator of migratory prey (such as Atlantic mackerel and Atlantic herring Clupea harengus). The biotic and/or abiotic conditions of northern waters may represent important limiting factors for the completion of the life cycle of this parasite in this geographical area. Among the drivers of the geographical range of Anisakis spp., certain abiotic conditions (i.e. temperature or salinity) may be considered limiting factors to the dispersal and range expansion of this parasite species, probably impeding the egg survival or the first developmental stages of hatched larvae to succeed in finding a suitable host in some oceanographic areas.
The combined “matrix” of the many definitive, intermediate and paratenic hosts of Anisakis spp. comprises a complex heterogeneous and constantly shifting mosaic, potentially influencing the endoparasite population structure and demography at both local and wider levels. As is the case for many other pelagic marine organisms, cetaceans are highly mobile, their movements driven by prey availability and suitable conditions for their reproduction42,86,87,88. The long-range dispersal of certain cetacean species, along with the dispersal contribution provided by highly migratory fish hosts, may maintain high levels of gene flow between Anisakis populations throughout their distribution range. However, cetaceans often show cryptic population genetic structure over a small geographical range, much more than expected for such highly migratory marine mammals89. Some genetic sub-structuring has been found in both A. simplex (s.s.) and A. pegreffii, despite the general high gene flow values observed between populations of these species2. This observation is supported by the haplotype network analysis provided in the present study (Fig. 4), showing a certain geographic separation in A. simplex (s.s.) individuals, with a significant differentiation between the parasites collected from cetaceans of the Norwegian Sea compared to conspecifics from definitive hosts of the NE Atlantic Ocean (on average, Fst ≈ 0.08) (Table 2). Cetacean population units with different feeding ecology90 and/or range of distribution may be responsible for maintaining the genetic differentiation observed in A. simplex (s.s.). Indeed, while a weakly significant genetic substructure of the parasite species appears to exist among metapopulations sampled from several definitive hosts, this structure seems to be stronger when analysing the parasite's population by geographic area of the host species. In turn, this seems to suggest that the geographic origin of the host species could be major driver explaining the sub-structuring of the parasite populations.
Interestingly, in a previous population genetic study of A. simplex (s.s.) larvae in herring from several NE Atlantic fishing grounds based on mtDNA cox2, the haplotype diversity values were, on average, in the same range as those observed in the present study91. Furthermore, the population showing the highest differentiation was the one from the most northern area, specifically the Norwegian Sea91. As herring is an important food source for cetacean species, such as minke whales, killer whales, and humpback whales (Megaptera novaeangliae), it is reasonable to expect that the Norwegian A. simplex (s.s.) population detected in herring overlaps with the one identified here based on network and haplotype analysis conducted on the corresponding adult stage in cetacean predators.
Moving to the southernmost waters of the NE Atlantic, significant genetic differentiation was observed (on average Fst ≈ 0.05) between parasite populations of A. pegreffii from dolphins in the Mediterranean Sea and Iberian Atlantic waters investigated in this study. Significant genetic differentiation was reported between populations of the short-beaked common dolphin from the Mediterranean Sea and adjacent Atlantic waters (Galicia and Portugal), reflecting a differential distribution and habitat use for populations of this dolphin species92. This finding suggests that the populations sub-structuring of the endoparasites (i.e. A. pegreffii) along their distribution range may overlap with different cetacean populations from different European waters.
Regarding the population genetics of A. physeteris identified in sperm whales stranded along the coast of the southern Adriatic Sea and the Tyrrhenian Sea, the TCS analysis has shown a star-like shape with many haplotypes (Fig. 6), most of which are shared by all the nematodes sampled in the same host from different geographical areas. However, a weak but significant level (p = 0.03) of population substructuring (on average Fst ≈ 0.01) was found between the metapopulation of A. physeteris collected from sperm whales stranded on the coast of Vasto (animals which died on the Southern Adriatic Sea coast but which come from the deeper waters of the Ionian area of the Mediterranean Sea39,93) and those from the dwarf sperm whale stranded along the Tyrrhenian Sea coast. A consistent level of differentiation (on average, Fst ≈ 0.18, p = 0.003) was observed between this parasite species recovered from the sperm whale from Scottish waters (Atlantic Ocean) and those from the Mediterranean Sea (data not shown). Although these preliminary findings need to be validated by examining a higher number of individuals from the NE Atlantic, they appear to reflect the Mediterranean Sea and the Atlantic Ocean sub-structuring observed in the main host of A. physeteris, the sperm whale. In fact, the genetic analysis conducted on whales stranded on the coast of the Adriatic Sea in two separate events (Varano in 2009 and Vasto in 2014) showed that all the specimens belonged to a unique haplotype, similar to other individuals from the Mediterranean Sea39,93,94, but genetically distinct from the adjacent populations of the Atlantic Ocean95.
Overall, these observations suggest that the genetic structure, the phylogeography and the ecology of the definitive hosts species are likely to also shape the genetic structure of their anisakid nematodes, as has been recently observed in other nematode parasites of marine mammals, such as the pinniped parasite Uncinaria lucasi96.
Conclusions
Studies on the adult stage of nematode parasites of the genus Anisakis mostly rely on occasional stranding events of their definitive hosts, making sampling unpredictable and opportunistic. At the same time, the scientific data obtained are crucial to understand the biology, host specificity, and life cycle of the Anisakis species. In fact, the ecology, evolutionary history, geographical distribution, and migration of cetaceans are among the drivers defining their parasites’ geographic range and population genetic structure. Drivers of marine speciation are rarely obvious, because the distributions of marine organisms (and changes in these distributions) tend to be poorly documented, implying a high level of uncertainty, and also because the oceans are continuous environment with few geographical barriers. Therefore, we have little understanding of the nature and mechanisms of reproductive isolation in the marine environment that determine the host-parasite association between Anisakis spp. and their cetacean hosts. This study indicates a congruence between the geographic distribution of the adult stages of the Anisakis species in their final hosts and the larval stages in their fish hosts. While Anisakis spp. are generalists for paratenic/transport hosts, the findings on definitive host-parasite association at the adult stage suggests instead a strong preference by some Anisakis spp. for certain cetacean species or families. This host preference is likely to be driven by coevolutionary aspects and mediated by definitive host feeding ecology and geographic distribution. Novel data on genetic structuring, geographical distribution, and relative proportions of different parasites species in their hosts can help in unravelling the complex life cycles of these parasites. Alongside the influence of abiotic and biotic limiting factors, the intermediate, paratenic and definitive hosts, according to their ecological characteristics, all contribute to shape the current distributions of Anisakis spp. in the world’s oceans.
Methods
Cetacean parasites sampling data
Samples of Anisakis spp. were collected from 34 individuals belonging to 11 cetacean species stranded in locations along the Mediterranean Sea and NE Atlantic Ocean coast, between 2004 and 2019 (see Table 4 and Fig. 7). All cetacean individuals were necropsied following a standardised method97. All parasites were recovered from the stomach chambers of their hosts, usually during necropsy, and generally it was possible to obtain only a random subsample of the Anisakis spp. specimens infecting in the host. On a few occasions it was possible to count or estimate the total burden of parasites infecting the host.
In total, N = 478 anisakid nematodes were identified to genus level based on morphological characters reviewed by Mattiucci et al.3. Mature adults were separated from pre-adults, based on the visibility in the former of caudal papillae and spicules in males and eggs in females). The sampled nematodes were stored frozen at − 20 °C, or in 70% ethanol, for further molecular analyses.
Genetic identification of anisakid parasites
The nematode specimens were identified by sequence analysis of the mitochondrial cytochrome c oxidase II (mtDNA cox2) gene locus23,25. Specimens of A. pegreffii and A. simplex (s.s.) (N = 134), as inferred from the mitochondrial marker, collected from cetaceans stranded in sympatric areas for these two sibling species, were also identified using nuclear markers. In particular, sequence analysis of the diagnostic gene locus elongation factor EF1 α − 1 of nDNA26 and direct genotyping determination of the diagnostic nuclear metallopeptidase nas10 locus by ARMS-PCR27 were performed on these 134 specimens. In addition, sequence analysis of the same gene locus (nas 10 nDNA) was carried out on those individuals which showed a heterozygote pattern.
Total DNA was extracted from ~ 2 mg of tissue from each specimen by using the Quick-gDNA Miniprep Kit (ZYMO RESEARCH), following the manufacturer’s instructions. For sequencing of the cox2 gene, polymerase chain reaction (PCR) amplification was performed using the primers 211F (5′-TTTTCTAGTTATATAGATTGRTTTYAT-3′) and 210R (5′-CACCAACTCTTAAAATTA TC-3′)98. PCR was carried out according to the procedures described by Mattiucci et al.23.
The EF1 α − 1 nDNA gene was amplified using the primers EF-F (5′-TCCTCAAGCGTTGTTATCTGTT-3′) and EF-R (5′-AGTTTTGCCACTAGCGGTTCC-3′)26. PCR conditions and procedures followed those reported by Mattiucci et al.26. The EF1 α − 1 nDNA sequences were compared with those previously deposited in GenBank, at the diagnostic positions (i.e., 186 and 286), as previously detailed by Mattiucci et al.26.
For sequencing the nas10 gene, PCR amplification was performed using the primers nas10F (5′-GATGTTCCTGCAAGTGATTG-3′) and nas10R (5′-CGCTATTAAGAGAGGGATCG-3′)27. PCR was carried out according to the procedures provided by Palomba et al.27. A direct genotyping determination of the nuclear nas10 locus was performed by ARMS-PCR at the gene locus nas10 by the combined use of OUT-F1 (5′-TATGGCAAATATTATTATCGTA-3′), OUT-R1 (5′-TATTTCCGACAGCAAACAA-3′), INN-F1 (5′-GCATTGTACACTTCGTATATT-3′), INN-R1 (5′-ATTTCTYCAGCAATCGTAAG-3′), following the procedures reported in Palomba et al.27. The PCR products were separated by electrophoresis using agarose gel (1.5%) stained with GelRed; 3 µL of the amplification products were visualized. The distinct banding patterns were detected using ultraviolet transillumination27.
The sequences obtained at the mtDNA cox2 locus were aligned with other Anisakis spp. sequences obtained in previous studies and deposited in GenBank, using ClustalX v2.099. Alignments were manually edited and trimmed with BioEdit v7.0.5.3100, resulting in 580 characters and 9 taxa.
Phylogenetic analysis inferred from results for the mtDNA cox2 gene locus
A phylogenetic tree of the cox2 mtDNA gene locus was constructed using Bayesian Markov Chain Monte Carlo (MCMC) analysis in BEAST v1.10.4101. The best-fit substitution model was selected using the Akaike information criterion (AIC) as implemented in jModeltest v2.1.7.102. GTR + I + G substitution model, strict molecular clock103 and a Yule speciation process were used as the tree priors104, with default parameters. A run of 10 million iterations was completed, logging parameter values after every 10,000 iterations, and checking for stationarity and effective sample sizes (ESS) from all the available parameters (cut-off > 200) with Tracer v1.7105. The first 10% of trees were discarded as burn in, and the remaining 9000 were analysed and visualized using TreeAnnotator v.1.10.4101, and FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/), respectively. Toxocara canis (AP017701) and Ascaris suum (X54253) were used as outgroups.
Intraspecific genetic diversity of Anisakis spp. inferred from results at the mtDNA cox2 gene locus
The mean pairwise differences, among and within populations of A. simplex (s.s.), A. pegreffii and A. physeteris from the different cetacean samples were calculated from mtDNA cox2 sequences. Spatial analysis of molecular variance (AMOVA), conducted with ARLEQUIN version 3.5, with 1000 permutations, was applied to the genetic data sets obtained for the populations of A. simplex (s.s.), A. pegreffii and A. physeteris. The Fst106 values were estimated by ARLEQUIN V3.5107 at the intraspecific level, among and within samples of A. simplex (s.s.), A. pegreffii and A. physeteris from different sampling areas. The Fst value ranges from a maximum of 1, indicating complete differentiation among sequences of pre-defined populations of the same Anisakis species collected from different regions to a value equal to a minimum of 0, which indicates no differentiation among the populations. Pairwise comparisons of Fst (assuming that p < 0.05 indicates a significant difference) were based on 1000 permutations of the data matrix. Intraspecific population genetic diversity of A. simplex (s.s.), A. pegreffii and A. physeteris among sampling areas was estimate based on the following standard statistical parameters: number of haplotypes (Nh), number of unique haplotypes (Nuh), nucleotide diversity (π), haplotype diversity (Hd), average number of differences (K), and number of polymorphic sites (S), using DnaSP V5.10.01 (http://www.ub.edu/dnasp)108. Haplotype network construction was carried out using PopART V1.7 (http://popart.otago.ac.nz/index.shtml)109. The analysis was performed using the statistical parsimony procedure (95% parsimony connection limit), implemented in TCS V3.5.1.2110.
Data availability
All data generated or analyzed during this study are included in this published article. The DNA sequences of the Anisakis species identified were deposited in the public sequence repository GenBank (NCBI National Center for Biotechnology Information—https://www.ncbi.nlm.nih.gov/genbank) [accession numbers: OM142467-81, OM210033-42].
References
Kuhn, T., Cunze, S., Kochmann, J. & Klimpel, S. Environmental variables and definitive host distribution: A habitat suitability modelling for endohelminth parasites in the marine realm. Sci. Rep. 6, 30246 (2016).
Mattiucci, S. & Nascetti, G. Advances and trends in the molecular systematics of anisakid nematodes, with implications for their evolutionary ecology and host-parasite co-evolutionary processes. Adv. Parasitol. 66, 47–148 (2008).
Mattiucci, S., Cipriani, P., Levsen, A., Paoletti, M. & Nascetti, G. Molecular epidemiology of Anisakis and Anisakiasis: An ecological and evolutionary road map. Adv. Parasitol. 99, 93–263 (2018).
Colón-Llavina, M. M. et al. Additional records of metazoan parasites from Caribbean marine mammals, including genetically identified anisakid nematodes. Parasitol. Res. 105, 1239–1252 (2009).
Iñiguez, A. M., Santos, C. P. & Vicente, A. C. P. Genetic characterization of Anisakis typica and Anisakis physeteris from marine mammals and fish from the Atlantic Ocean off Brazil. Vet. Parasitol. 165, 350–356 (2009).
Gomes, T. L. et al. Anisakis spp. in toothed and baleen whales from Japanese waters with notes on their potential role as biological tags. Parasitol. Int. 80, 102228 (2021).
Irigoitia, M. et al. Genetic identification of Anisakis spp. (Nematoda: Anisakidae) from cetaceans of the Southwestern Atlantic Ocean: Ecological and zoogeographical implications. Parasit. Res. 120, 1–13 (2021).
Ugland, K. I., Strømnes, E., Berland, B. & Aspholm, P. E. Growth, fecundity and sex ratio of adult whaleworm (Anisakis simplex; Nematoda, Ascaridoidea, Anisakidae) in three whale species from the North-East Atlantic. Parasitol. Res. 92, 484–489 (2004).
Berland, B. Musings on nematode parasites. Fisken og Havet 11, 1–26 (2006).
Roca-Geronès, X., Alcover, M. M., Godínez-González, C., Montoliu, I. & Fisa, R. Hybrid genotype of Anisakis simplex (s.s.) and A. pegreffii identified in third- and fourth-stage larvae from sympatric and allopatric Spanish marine waters. Animals 11, 2458 (2021).
Smith, J. Ulcers associated with larval Anisakis simplex B (Nematoda: Ascaridoidea) in the forestomach of harbour porpoises Phocoena phocoena (L.). Can. J. Zool. 67, 2270–2276 (1989).
Abollo, E., Lopez, A., Gestal, C., Benavente, P. & Pascual, S. Macroparasites in cetaceans stranded on the northwestern Spanish Atlantic coast. Dis. Aquat. Org. 32, 227–231 (1998).
Hrabar, J., Bočina, I., Gudan Kurilj, A., Đuras, M. & Mladineo, I. Gastric lesions in dolphins stranded along the Eastern Adriatic coast. Dis. Aquat. Organ. 125, 125–139 (2017).
Pons-Bordas, C. et al. Recent increase of ulcerative lesions caused by Anisakis spp. in cetaceans from the north-east Atlantic. J. Helminthol. 94, E127 (2020).
Ryeng, K. A., Lakemeyer, J., Roller, M., Wohlsein, P. & Sieber, U. Pathological findings in bycaught harbour porpoises (Phocoena phocoena) from the coast of Northern Norway. Polar Biol. 45, 45–57 (2021).
Mattiucci, S., Cipriani, P., Paoletti, M., Levsen, A. & Nascetti, G. Reviewing biodiversity and epidemiological aspects of anisakid nematodes from the North East Atlantic Ocean. J. Helminthol. 91, 422–439 (2017).
Mattiucci, S. et al. Novel polymorphic microsatellite loci in Anisakis pegreffii and A. simplex (s.s.) (Nematoda: Anisakidae): Implications for species recognition and population genetic analysis. Parasitology 146, 1387–1403 (2019).
Shamsi, S., Sprohnle-Barrera, C. & Hossen, M. D. S. Occurrence of Anisakis spp. (Nematoda: Anisakidae) in a pygmy sperm whale Kogia breviceps (Cetacea: Kogiidae) in Australian waters. Dis. Aquat. Organ. 134, 65–74 (2019).
Cavallero, S., Nadler, S. A., Paggi, L., Barros, N. B. & D’Amelio, S. Molecular characterization and phylogeny of anisakid nematodes from cetaceans from southeastern Atlantic coasts of USA, Gulf of Mexico, and Caribbean Sea. Parasitol. Res. 108, 781–792 (2011).
Klimpel, S. & Palm, H. W. Anisakid nematode (Ascaridoidea) life cycles and distribution: increasing zoonotic potential in the time of climate change? In Progress in Parasitology, Parasitology Research Monographs Vol. 2 (ed. Mehlhorn, H.) 201–222 (Springer, 2011).
Li, L. et al. Molecular phylogeny and dating reveal a terrestrial origin in the early Carboniferous for Ascaridoid nematodes. Syst. Biol. 67, 888–900 (2018).
Shamsi, S. Recent advances in our knowledge of Australian anisakid nematodes. Int. J. Parasitol. Parasites Wildl. 3, 178–187 (2014).
Mattiucci, S. et al. Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi n. sp. for A. simplex sp. C (Nematoda: Anisakidae). J. Parasitol. 100, 199–214 (2014).
D’Amelio, S. et al. Genetic markers in ribosomal DNA for the identification of members of the genus Anisakis (Nematoda: Ascaridoidea) defined by polymerase-chain-reaction-based restriction fragment length polymorphism. Int. J. Parasitol. 30, 223–226 (2000).
Valentini, A. et al. Genetic relationships among Anisakis species (Nematoda: Anisakidae) inferred from mitochondrial cox2 sequences, and comparison with allozyme data. J. Parasitol. 92, 156–166 (2006).
Mattiucci, S. et al. No more time to stay ‘single’ in the detection of Anisakis pegreffii, A. simplex (s.s.) and hybridization events between them: A multi-marker nuclear genotyping approach. Parasitology 143, 998–1011 (2016).
Palomba, M., Paoletti, M., Webb, S. C., Nascetti, G. & Mattiucci, S. A novel nuclear marker and development of an ARMS-PCR assay targeting the metallopeptidase 10 (nas 10) locus to identify the species of the Anisakis simplex (s. l.) complex (Nematoda, Anisakidae). Parasite 27, 39 (2020).
Mladineo, I. et al. Anisakis simplex complex: Ecological significance of recombinant genotypes in an allopatric area of the Adriatic Sea inferred by genome-derived simple sequence repeats. Int. J. Parasitol. 47, 215–223 (2017).
Bello, E., Paoletti, M., Webb, S. C., Nascetti, G. & Mattiucci, S. Cross-species utility of microsatellite loci for the genetic characterisation of Anisakis berlandi (Nematoda: Anisakidae). Parasite 27, 9 (2020).
Bello, E. et al. Investigating the genetic structure of the parasites Anisakis pegreffii and A. berlandi (Nematoda: Anisakidae) in a sympatric area of the southern Pacific Ocean waters using a multilocus genotyping approach: First evidence of their interspecific hybridization. Infect. Genet. Evol. 92, 104887 (2021).
Klapper, R. et al. Anisakid nematodes in beaked redfish (Sebastes mentella) from three fishing grounds in the North Atlantic, with special notes on distribution in the fish musculature. Vet. Parasit. 207, 72–80 (2015).
Bušelić, I. et al. Geographic and host size variations as indicators of Anisakis pegreffii infection in European pilchard (Sardina pilchardus) from the Mediterranean Sea: Food safety implications. Int. J. Food Microb. 266, 126–132 (2018).
Cipriani, P. et al. Anisakis pegreffii (Nematoda: Anisakidae) in European anchovy Engraulis encrasicolus from the Mediterranean Sea: Fishing ground as a predictor of parasite distribution. Fish. Res. 202, 59–68 (2018).
Cipriani, P. et al. The Mediterranean European hake, Merluccius merluccius: Detecting drivers influencing the Anisakis spp. larvae distribution. Fish. Res. 202, 79–89 (2018).
Levsen, A. et al. A survey of zoonotic nematodes of commercial key fish species from major European fishing grounds—Introducing the FP7 PARASITE exposure assessment study. Fish. Res. 202, 4–21 (2018).
Gibson, D. I. et al. A survey of the helminth parasites of cetaceans stranded on the coast of England and Wales during the period 1990–1994. J. Zool. 244, 563–574 (1998).
Mattiucci, S. et al. Evidence for a new species of Anisakis Dujardin, 1845: Morphological description and genetic relationships between congeners (Nematoda: Anisakidae). Syst. Parasitol. 61, 157–171 (2005).
Blažeković, K., Pleić, I. L., Đuras, M., Gomerčić, T. & Mladineo, I. Three Anisakis spp. isolated from toothed whales stranded along the eastern Adriatic Sea coast. Int. J. Parasitol. 45, 17–31 (2015).
Mazzariol, S. et al. Multidisciplinary studies on a sick-leader syndrome-associated mass stranding of sperm whales (Physeter macrocephalus) along the Adriatic coast of Italy. Sci. Rep. 8, 11577 (2018).
Gomerčić, M. et al. Bottlenose dolphin (Tursiops truncatus) depredation resulting in larynx strangulation with gill-net parts. Mar. Mammal Sci. 25, 392–401 (2009).
Pyenson, N. The high fidelity of the cetacean stranding record: Insights into measuring diversity by integrating taphonomy and macroecology. Proc. R. Soc. B. 278, 3608–3616 (2011).
MacLeod, C. D., Santos, B., López Fernandez, A. & Pierce, G. Relative prey size consumption in toothed whales: Implications for prey selection and level of specialisation. Mar. Ecol. Prog. Ser. 326, 295–307 (2006).
Santos, M. B. et al. Pygmy sperm whales Kogia Breviceps in the Northeast Atlantic: New information on stomach contents and strandings. Mar. Mammal Sci. 22, 600–616 (2006).
Covelo, P., Martínez-Cedeira, J., Llavona, A., Díaz, J. & López Fernandez, A. Strandings of Beaked Whales (Ziphiidae) in Galicia (NW Spain) between 1990 and 2013. J. Mar. Biol. Assoc. U. K. 1, 1–7 (2016).
Moura, J. et al. Stranding events of Kogia whales along the Brazilian Coast. PLoS ONE 11, e0146108 (2016).
Cordes, D. O. The causes of whale strandings. N. Z. Vet. J. 30, 21–24 (1982).
Frantzis, A. Does acoustic testing strand whales?. Nature 392, 29 (1998).
Laist, D. W., Knowlton, A. R., Mead, J. G., Collet, A. S. & Podesta, M. Collisions between ships and whales. Mar. Mammal Sci. 17, 35–75 (2001).
Jepson, P. D. et al. Gas-bubble lesions in stranded cetaceans. Nature 425, 575–576 (2003).
Pierce, G. J., Santos, M. B., Smeenk, C., Saveliev, A. & Zuur, A. F. Historical trends in the incidence of strandings of sperm whales (Physeter macrocephalus) on North Sea coasts: An association with positive temperature anomalies. Fish. Res. 87, 219–228 (2007).
Coombs, E. et al. What can cetacean stranding records tell us? A study of UK and Irish cetacean diversity over the past 100 years. Mar. Mammal Sci. 35, 1527–1555 (2019).
Fossi, M. C., Baini, M., Panti, C. & Baulch, S. Chapter 6—Impacts of marine litter on cetaceans: A focus on plastic pollution. In Marine Mammal Ecotoxicology (eds Fossi, M. C. & Panti, C.) 147–184 (Academic Press, 2018).
Alexiadou, P., Foskolos, I. & Frantzis, A. Ingestion of macroplastics by odontocetes of the Greek Seas, Eastern Mediterranean: Often deadly!. Mar. Poll. Bull. 146, 67–75 (2019).
Nicol, C. et al. Anthropogenic threats to Wild Cetacean welfare and a tool to inform policy in this area. Vet. Sci. Res. J. 7, 57 (2020).
Abollo, E., Paggi, L., Pascual, S. & D’Amelio, S. Occurrence of recombinant genotypes of Anisakis simplex s.s. and Anisakis pegreffii (Nematoda: Anisakidae) in an area of sympatry. Infect. Genet. Evol. 3, 175–181 (2003).
Marques, J. F., Cabral, H., Busi, M. & D’Amelio, S. Molecular identification of Anisakis species from Pleuronectiformes off the Portuguese coast. J. Helminthol. 80, 47–51 (2006).
Lee, M. H., Cheon, D. & Choi, C. Molecular genotyping of Anisakis species from Korean sea fish by polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP). Food Control 20, 623–626 (2009).
Suzuki, J., Murata, R., Hosaka, M. & Araki, J. Risk factors for human Anisakis infection and association between the geographic origins of Scomber japonicus and anisakid nematodes. Int. J. Food Microbiol. 137, 88–93 (2010).
Molina-Fernández, D. et al. Fishing area and fish size as risk factors of Anisakis infection in sardines (Sardina pilchardus) from Iberian waters, southwestern Europe. Int. J. Food Microb. 203, 27–34 (2015).
Cipriani, P. et al. Genetic identification and distribution of the parasitic larvae of Anisakis pegreffii and Anisakis simplex (s.s.) in European hake Merluccius merluccius from the Tyrrhenian Sea and Spanish Atlantic coast: Implications for food safety. Int. J. Food Microbiol. 198, 1–8 (2015).
Gómez-Mateos, M., Merino-Espinosa, G., Corpas-López, V., Valero-López, A. & Martín-Sánchez, J. A multi-restriction fragment length polymorphism genotyping approach including the beta-tubulin gene as a new differential nuclear marker for the recognition of the cryptic species Anisakis simplex s.s. and Anisakis pegreffii and their hybridization events. Vet. Parasitol. 283, 109162 (2020).
Klimpel, S., Busch, M. W., Kuhn, T., Rohde, A. & Palm, H. The Anisakis simplex complex off the South Shetland Islands (Antarctica): Endemic populations versus introduction through migratory hosts. Mar. Ecol. Progr. Ser. 40, 1–11 (2010).
Santoro, M. et al. Helminth parasites of the dwarf sperm whale Kogia sima (Cetacea: Kogiidae) from the Mediterranean Sea, with implications on host ecology. Dis. Aquat. Organ. 129, 175–182 (2018).
Mattiucci, S., Nascetti, G., Bullini, L., Orecchia, P. & Paggi, L. Genetic structure of Anisakis physeteris and its differentiation from the Anisakis simplex complex (Ascaridida: Anisakidae). Parasitology 93, 383–387 (1986).
Palomba, M., Mattiucci, S., Crocetta, F., Osca, D. & Santoro, M. Insights into the role of deep-sea squids of the genus Histioteuthis (Histioteuthidae) in the life cycle of ascaridoid parasites in the Central Mediterranean Sea waters. Sci. Rep. 11, 7135 (2021).
Clarke, M. R., Martins, H. R. & Pascoe, P. The diet of sperm whales (Physeter macrocephalus Linnaeus 1758) off the Azores. Philos. Trans. R. Soc. Lond. B. 339, 67–82 (1993).
Santos, M. & Pierce, G. A note on niche overlap in teuthophagous whales in the northern Northeast Atlantic. Phuket Mar. Biol. Cent. Res. Bull. 66, 291–298 (2005).
Rendell, L. & Frantzis, A. Mediterranean Sperm Whales, Physeter macrocephalus: The precarious state of a lost tribe. In Advances in Marine Biology (eds Notarbartolo di Sciara, G. et al.) 37–74 (Academic Press, 2016).
Foskolos, I., Koutouzi, N., Polychronidis, L., Alexiadou, P. & Frantzis, A. A taste for squid: the diet of sperm whales stranded in Greece, Eastern Mediterranean. Deep Sea Res. I Oceanogr. Res. Pap. 155, 103164 (2020).
Mattiucci, S. et al. Genetic heterogeneity within Anisakis physeteris (sensu lato) (Nematoda: Anisakidae) from sperm whales, Physeter macrocephalus, from Mediterranean Sea (Apulian coast) and Atlantic Ocean (Canaries coast). Abstract of XXVI Congresso Nazionale SoIPa. Parassitologia 52, 357 (2010).
Mattiucci, S. et al. Genetic identification and insights into the ecology of Contracaecum rudolphii A and C. rudolphii B (Nematoda: Anisakidae) from cormorants and fish of aquatic ecosystems of Central Italy. Parasitol. Res. 119, 1243–1257 (2020).
Karvonen, A., Jokela, J. & Laine, A. L. Importance of sequence and timing in parasite coinfections. Trends Parasitol. 35, 109–118 (2019).
Paggi, L. et al. A new species of Anisakis Dujardin, 1845 (Nematoda: Anisakidae) from beaked whale (Ziphiidae): Allozyme and morphological evidence. Syst. Parasitol. 40, 161–174 (1998).
Mattiucci, S., Paoletti, M. & Webb, S. C. Anisakis nascettii n. sp. (Nematoda: Anisakidae) from beaked whales of the southern hemisphere: Morphological description, genetic relationships between congeners and ecological data. Syst. Parasitol. 74, 199–217 (2009).
Leatherwood, S. & Reeves, R. R. The Sierra Club Handbook of Whales and Dolphins 302 (Sierra Club Books, 1983).
Ross, G. J. B. The smaller cetaceans of the South East coast of southern Africa. Ann. Cape Prov. Mus. Nat. Hist. 15, 173–410 (1984).
Santos, B. et al. Feeding ecology of Cuvier’s beaked whale (Ziphius cavirostris): A review with new information on the diet of this species. J. Mar. Biol. Assoc. U. K. 81, 687–694 (2001).
Lakemeyer, J. et al. Anisakid nematode species identification in harbour porpoises (Phocoena phocoena) from the North Sea, Baltic Sea and North Atlantic using RFLP analysis. Int. J. Parasitol. Parasites Wildl. 12, 93–98 (2020).
Højgaard, D. No significant development of Anisakis simplex (Nematoda, Anisakidae) eggs in the intestine of long-finned pilot whales, Globicephala melas (Traill, 1809). Sarsia 84, 479–482 (1999).
Smith, J. W. & Wootten, R. Experimental studies on the migration of Anisakis sp. larvae (Nematoda: ascaridida) into the flesh of herring, Clupea harengus L. Int. J. Parasitol. 5, 133–136 (1975).
Iglesias, L., Valero, A., Benítez, R. & Adroher, F. J. In vitro cultivation of Anisakis simplex: Pepsin increases survival and moulting from fourth larval to adult stage. Parasitology 123, 285–291 (2001).
Mladineo, I. & Poljak, V. Ecology and genetic structure of zoonotic Anisakis spp. from adriatic commercial fish species. Appl. Environ. Microbiol. 80, 1281–1290 (2014).
Mladineo, I., Bušelić, I., Hrabar, J., Vrbatović, A. & Radonić, I. Population parameters and mito-nuclear mosaicism of Anisakis spp. in the Adriatic Sea. Mol. Biochem. Parasitol. 212, 46–54 (2017).
Levsen, A. et al. Anisakis species composition and infection characteristics in Atlantic mackerel, Scomber scombrus, from major European fishing grounds—Reflecting changing fish host distribution and migration pattern. Fish. Res. 202, 112–121 (2018).
Gay, M. et al. Infection levels and species diversity of ascaridoid nematodes in Atlantic cod, Gadus morhua, are correlated with geographic area and fish size. Fish. Res. 202, 90–102 (2018).
Stevick, P. et al. Segregation of migration by feeding ground origin in North Atlantic humpback whales (Megaptera novaeangliae). J. Zool. 259, 231–237 (2003).
Lambert, E. et al. Cetacean range and climate in the eastern North Atlantic: Future predictions and implications for conservation. Glob. Change Biol. 20, 1782–1793 (2014).
Szesciorka, A. et al. Timing is everything: Drivers of interannual variability in blue whale migration. Sci. Rep. 10, 7710 (2020).
Hoelzel, A. R., Goldsworthy, S. D. & Fleischer, R. C. Population genetic structure. In Marine Mammal Biology: An Evolutionary Approach (ed. Hoelzel, A. R.) 1–134 (Blackwell Publishing, 2002).
Lahaye, V. et al. Long-term dietary segregation of common dolphins Delphinus delphis in the Bay of Biscay, determined using cadmium as an ecological tracer. Mar. Ecol. Prog. Ser. 305, 275–285 (2005).
Mattiucci, S. et al. Population genetic structure of the parasite Anisakis simplex (s.s.) collected in Clupea harengus L. from North East Atlantic fishing grounds. Fish. Res. 202, 103–111 (2018).
Natoli, A. et al. Conservation genetics of the short-beaked common dolphin (Delphinus delphis) in the Mediterranean Sea and in the eastern North Atlantic Ocean. Conserv. Genet. 9, 1479–1487 (2008).
Mazzariol, S. et al. Sometimes sperm whales (Physeter macrocephalus) cannot find their way back to the high seas: A multidisciplinary study on a mass stranding. PLoS ONE 6, e19417 (2011).
Mazzariol, S. et al. Dolphin Morbillivirus associated with a mass stranding of sperm Whales, Italy. Emerg. Infect. Dis. 23, 144–146 (2017).
Podestà, M. et al. Cuvier’s beaked whale, Ziphius cavirostris, distribution and occurrence in the Mediterranean Sea: High-use areas and conservation threats. Adv. Mar. Biol. 75, 103–140 (2016).
Davies, K., Pagan, C. & Nadler, S. A. Host population expansion and the genetic architecture of the pinniped hookworm Uncinaria lucasi. J. Parasitol. 106, 383–391 (2020).
IJsseldijk, L. L., Brownlow, A. C. & Mazzariol, S. European best practice on cetacean post-mortem investigation and tissue sampling (ed. IJsseldijk, L. L., Brownlow, A. C., & Mazzariol, S.) 1–72 (ASCOBANS/ACCOBAMS, 2019).
Nadler, S. A. & Hudspeth, D. S. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: Hypotheses of structural and sequence evolution. J. Parasitol. 86, 380–393 (2000).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Hall, T. A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999).
Suchard, M. A. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Zuckerkandl, E. & Pauling, L. Molecular disease, evolution, and genetic heterogeneity. In Horizons in Biochemistry (eds Kasha, M. & Pullman, B.) 189–225 (Academic Press, 1962).
Gernhard, T. The conditioned reconstructed process. J. Theor. Biol. 253, 769–778 (2008).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Weir, B. & Cockerham, C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Librado, P. & Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Bandelt, H., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Clement, M., Posada, D. & Crandall, K. A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659 (2000).
Acknowledgements
We affectionately dedicate this paper to the memory of our colleague and friend Professor Dario Angeletti from the Department of Ecological and Biological Sciences (Tuscia University) who tragically left us on December, the 7th, 2021, during the final stages of writing this paper. He was the director of the Tuscia University unit of Centro Interuniversitario di Ricerca sui Cetacei (CIRCE), and a researcher with a great fondness for cetaceans of the Mediterranean Sea. The authors are very grateful to all the researchers and collaborators who worked on the stranded cetaceans, providing the precious parasite specimens that allowed this research.
Funding
The study was supported by GRANT Ateneo Sapienza Grandi Progetti 2020 (Principal Investigator: S. Mattiucci). P. Cipriani and M. Palomba were funded by Italian MUR-PON grants. CESAM is supported by FCT/MCTES (UIDP/50017/2020 + UIDB/50017/2020 + LA/P/0094/2020). G.J. Pierce acknowledges support from the Spanish Ministerio de Ciencia e Innovacion (project “TRANSITION”, RTI2018-099607-B-I00). The Galician stranding network is supported by the regional government Xunta de Galicia/Dirección Xeneral de Patrimonio Natural, that also provided the legal permit for the collection of biological samples. AL is funded by national funds (OE), through FCT—Fundação para a Ciência e a Tecnologia, I.P., in the scope of the framework contract foreseen in the numbers 4, 5 and 6 of the article 23, of the Decree-Law 57/2016, of August 29, changed by Law 57/2017, of July 19. Pelagos Cetacean Research Institute wish to express their gratitude for the support of OceanCare (Switzerland) to all their research on Cuvier’s and sperm whales, including sampling and necropsies.
Author information
Authors and Affiliations
Contributions
P.Cip. and S.M. organized and designed the whole study, and wrote the main manuscript. M.P., L.G., M.P. worked on the genetic identification of the parasites and the genetic data analyses, plus edited tables and figures. F.M., S.M., M.S., R.A., P.Cov., A.L., B.S., G.P., A.B., N.D., B.M., A.F., V.A., D.H., B.M., A.L., G.N. worked on cetacean postmortem inspection, collected parasite samples, and all participated to the revision process of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cipriani, P., Palomba, M., Giulietti, L. et al. Distribution and genetic diversity of Anisakis spp. in cetaceans from the Northeast Atlantic Ocean and the Mediterranean Sea. Sci Rep 12, 13664 (2022). https://doi.org/10.1038/s41598-022-17710-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-17710-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.