Abstract
Cysteine-rich receptor-like-kinases (CRKs), a transmembrane subfamily of receptor-like kinase, play crucial roles in plant adaptation. As such cotton is the major source of fiber for the textile industry, but environmental stresses are limiting its growth and production. Here, we have performed a deep computational analysis of CRKs in five Gossypium species, including G. arboreum (60 genes), G. raimondii (74 genes), G. herbaceum (65 genes), G. hirsutum (118 genes), and G. barbadense (120 genes). All identified CRKs were classified into 11 major classes and 43 subclasses with the finding of several novel CRK-associated domains including ALMT, FUSC_2, Cript, FYVE, and Pkinase. Of these, DUF26_DUF26_Pkinase_Tyr was common and had elevated expression under different biotic and abiotic stresses. Moreover, the 35 land plants comparison identified several new CRKs domain-architectures. Likewise, several SNPs and InDels were observed in CLCuD resistant G. hirsutum. The miRNA target side prediction and their expression profiling in different tissues predicted miR172 as a major CRK regulating miR. The expression profiling of CRKs identified multiple clusters with co-expression under certain stress conditions. The expression analysis under CLCuD highlighted the role of GhCRK057, GhCRK059, GhCRK058, and GhCRK081 in resistant accession. Overall, these results provided primary data for future potential functional analysis as well as a reference study for other agronomically important crops.
Similar content being viewed by others
Introduction
In nature, plants are exposed to diverse environmental stresses, including biotic and abiotic stresses. To defend against these stresses, plants have evolved layered immune systems. This includes patterns-triggered immunity (PTI)1 that is induced by pattern recognition receptors (PRRs)2. Whereas effector-triggered immune (ETI) is activated when plants detect pathogens' RNAs and proteins-based effector molecules. From pathogens’ perspective, these effectors suppress both PTI and ETI and establish effector-triggered susceptibility (ETS)3,4,5. The plasma membrane possesses embedded proteins with extracellular and intracellular domains, including receptor-like kinases (RLKs) and receptor-like proteins (RLPs)6 that generally participate in PTI. The extracellular domain involves host–pathogen protein–protein interaction and signal perception, while the intracellular kinase domains transduce that signal and activate signaling pathways7,8,9,10,11,12. The RLKs have a potential role in different signaling mechanisms, including stress responses, hormone regulation, and plant growth and development13,14. RLKs and RLPs involve in regulations of several cellular mechanisms to strengthen plant adaptation under different environmental stresses. Multiple genome-wide studies in plants have identified RLKs and RLPs, but fewer have been biochemically and functionally characterized. The evolutionary divergence and speciation have been triggered for subfunctionalization and neofunctionalization of proteins, including RLKs and RLPs.
Cysteine-rich receptor-like kinase (CRKs), harboring Domain Unknown Function 26 (DUF 26; Gnk2 or Stress-antifungal) domain, is an extracellular domain that consists of the conserved cysteine-rich motif (C-X8-C-X2-C) in its core and possesses antifungal and salt-stress responsive activities. Thus far, the best-characterized CRKs identified in Gingko biloba consists of a single DUF26, which acts as mannose-binding lectin and provides resistance against the fungal pathogen15. The structural analysis of Arabidopsis PDLP5 and PDLP8 ectodomains is also similar to fungal lectins but in plants, it has an additional domain for carbohydrate-binding16. In Arabidopsis, the AtCRKs are transcriptionally induced under abiotic stresses such as salt, drought, UV light, heat, salicylic acid17,18,19,20,21. In addition, a subset of CRKs is strongly induced in response to pathogens and pathogen-mimic stimuli19,20. Similarly, the overexpression of Arabidopsis CRK4, CRK5, CRK6, CRK13, and CRK36 exhibited enhanced resistance to a bacterial pathogen Pseudomonas syringae and activated both early and late PTI responses17,21,22. The CRKs are categorized into three subgroups including cysteine-rich receptor-like secreted proteins (CRRSPs; single peptide followed by DUF26), cysteine-rich receptor-like protein kinases (CRKs; single peptide, two DUF26 domains, one transmembrane domain, and one kinase domain), plasmodesmata localized proteins (PDPs; with single DUF26 domain). These are involved in pathogen response, intra signaling, systematic signaling, and viral movement target23. A recent study identified CRKs in 32 plant species and algae genomes and classified them into nine subclasses i.e., sdCRRSP, ddCRRSP, PDLP, sdCRK, CRK_I, CRK_II, tdCRK, qdCRK, and qdCRRSP15. While the essential roles in plant adaptation are documented, their functions in Gossypium sp. are not explored.
The Gossypium genus encompasses 54 species with 47 diploids (2n = 26), and seven tetraploids (2n = 4x = 52)24. Among these species, only four are widely cultivated globally for fiber production. This includes two tetraploids (G. hirsutum; AtDt1 and G. barbadense; AtDt2) and two diploids (G. arboreum; A2, G. herbaceum; A1) species. According to the cotton polyploidization theory, the tetraploid AADD genome originated due to polyploidization of the A-like genome and D-like genome25,26,27,28,29,30. It is reported that the diploid species are resistant to several viral and fungal diseases as compared to allotetraploid28,31,32. Thus, a comparative study among resistant and susceptible species are essential to understand plant resistance mechanism for developing resistant cultivars. The current study comprises of genomic, transcriptomic, proteomic, and miRNA target site prediction study of CRK genes among five species, including G. arboreum, G. raimondii, G. herbaceum, G. barbadense, and G. hirsutum. Findings of this study have provided comprehensive insight into the CRKs’ evolution, expression patterns, interaction with viral proteins, genetic diversity of resistant and susceptible accessions, and miRNA target site predictions in Gossypium sp.
Material and methods
Identification and classification
The complete genome of G. hirsutum (Ghir: HAU_v1/v1.1), G. barbadense (Gbar; genome HAU_v2_a1), G. herbaceum (Gher; A1-0076_WHUv3.0), G. raimondii (Gra; BGI-CGP_v1.0), and G. arboreum (Gar; CRI-A2_v1.0_a1.0) and their associated data were retrieved from Cottongen and CottonFGD databases25,27,28,33,34,35. These protein sequences were scanned through the Pfam database in the local server using the Pfam-Scan tool36 with default parameters. All genes having DUF 26 (PF01657; Gnk2 or Stress-antifungal) domains were considered as CRKs. The identified proteins sequence was scanned with the Inter-Pro database and filtered with IPR038408 and IPR002902 accessions for further validation. In addition, we have also mapped different available genome assemblies to make them more applicable for more than one assemblies of the same species. For instance, we have mapped Gh_HAU_v1 / v1.1 (Ghir_A11G008640) with Gh_CRI_v1 (Gh_A11G085800.1) and Ghir_ BGI_v1 (CotAD_01546). Similarly, Gra_D5_B CGP_v1.0 (Cotton_D_gene_10022874) with Gra_JGI_v2.0 (Gorai.001G109400)25,27,28,33,34,35.
Conserved domain architecture was carried out to find duplicated domains and additional associated domains with stress-antifungal motifs37 protocol. The predicted domains were arranged at their specific site on amino acid sequence using the Perl program. Three different classification methods were implemented in this study; (1) types and location (N-terminal or C-terminal) of additional domains associated with the Stress-antifungal/DUF 206/Gnk2 domain with irrespective of duplicated domains, (2) complete domains, and (3) literature classifications e.g. sdCRRSP, ddCRRSP, PDLP, sdCRK, tdCRK and qdCRK15. We also included 35 land plants including mosses, bryophyte, gymnosperm, and angiosperm for evolutionary study of CRKs.
Protein statistics, chromosomal mapping, intron–exon distribution, and motif analysis
All gene and proteins associated data were retrieved from Cottongen, including protein length, molecular weight (kDa), charges, grand average of hydropathy, isoelectric point (Ip), chromosome start, and end. The chromosomal mapping was carried with TBTools gene location, intron–exon distribution generated with gene display server38, structural and functional motifs were detected with the MEME motif, and PROSIT Motifs discovery server39,40.
Evolution and diversity analysis of CRKs in Gossypium
An advanced comparative genomics tool, OrthoFinder41 was exploited to understand evolution and diversity in CRK proteins among five species. An additional DIAMOND tool was used for fast sequence similarity searches42. The graph clustering was done with the MCL clustering algorithm43. The gene tree inference and a distance matrix of the orthogroups were constructed with DendroBLAST44. A distance-based phylogeny tree was constructed using FastME 2.045. For multiple sequence alignment, MAFTT 7.0 was used46. The maximum likelihood phylogenetic tree of large alignment was constructed using FastTreeMP47. To construct the Circos plot of five genomes, a BlastP program was used to determine the linkage and the circular plot was constructed with Advance Circos plot packages in TBTools.
Expression profiling of CRKs genes
The expression profiling data is divided into three categories, i.e., tissue-specific (leaf, stem, root, ovule, etc.) expression, abiotic stress-specific (cold, heat, salinity, drought) expression, and biotic stress-specific CLCuD (Cotton leaf curl virus disease). To determine the expression profiling, the publicly available RNA-seq (SRP044705, SRP042128, SRP017168, SRP001603, SRP009820, and SRP027533) at CottonFGD48, whitefly infestation on CLCuD susceptible accession of G. hirsutum (SAMN07519654, SAMN07519653, SAMN07519652, SAMN07519651, SAMN07519650, SAMN07519649)49 and whitefly infestation on CLCuD resistant accession of G. hirsutum (SAMN07251316 and SAMN07251315)50 were used. The transcript level was calculated in fragments per kilo base per million (FPKM) by RNA-seq data pipelines. The gene expression clustering was carried using TBTools with parameters; log2 base, column cluster, and row cluster.
Protein–protein interaction network and host–pathogen model docking
The CRKs protein–protein interaction network was generated using a STRING server with G. raimondii proteome interaction background. Begomovirus, a genus of the Geminiviridae family, also known as plant virus, infects a wide range of dicotyledonous plants. In cotton plants, it causes CLCuD. So, we included all CLCuD viral proteins (AV1, AV2, AC3, AC2, AC1, AC4, and C5) for their possible interaction with cotton CRKs. The sequence-based interaction was predicted using the Host–Pathogen Interaction predictor (HOPITOR)51. The 3D structure of CRKs and viral proteins were predicted using the I-TASSER server52. The host–pathogen protein docking was carried with ZDOCK53. The protein complex was visualized with discovery stadio54 and the active sites and interactive bonds were presented with Ligplot + 55.
SNPs and InDels determination in CLCuD resistant and susceptible G. hirsutum accession
To find the genetic diversity of CRKs in cotton leaf curl disease-resistant and susceptible G. hirsutum accessions, a genome resequencing data of Mac7 (CLCuD resistant accession; developed by USDA through the breeding program) and Coker 312 (highly susceptible to CLCuD accession) was used. The resequencing data NCBI: PRJNA756435 (Mac7) and NCBI: PRJNA542238 (for Coker 312) has been used to find SNPs and InDels in the CRK genes. The raw reads of Mac7 and Coker 312 mapped to TM_1 reference gnome (HAU-AD1_genome_v1.0_v1.1) using a BWA aligner and followed the next-generation sequencing pipeline similar to Zhao et al.56 to find variant calling format files of CRK genes. The identified SNPs and InDels were annotated using the SnpEff tool with the reference genome (HAU-AD1_genome_v1.0_v1.1)26.
Target site prediction and expression profiling of miRNA
To find the miRNA target site in CRK coding sequences of Gossypium sp. mature miRNA sequences were retrieved from the Plant non-coding RNA database57 and PmiREN58. These downloaded miRNA and CDS sequences of Gossypium sp. were used as input data in psRNA target: a plant small RNA target analysis59.
The expression profiling of CRK-targeted miRNA was assessed using the miRNA-seq of Gossypium sp. data located at PmiREN (Plant Micro RNA Encyclopedia )58 covering different tissues including anther, fiber, embryogenic, hypocotyl, leaf, ovule, root, shoot apical, stem, and apexes.
Plant growth and CLCuD stress
To validate the RNA-seq data of GhCRKs, we selected two G. hirsutum accessions, Mac7 (resistant to CLCuD) and Coker 312 (susceptible to CLCuD). A set of 20 plants was sown in the glasshouse for each accession. After five weeks of post-germination, one set of each accession (10 plants) was transplanted in the net house to expose the whitefly (CLCuD career vector). After two weeks of post-transplantation, a high population of whitefly was seen on Coker 312’s as well as Mac7 plants. All Coker 312 were 100% infected with severe symptoms, while no symptom was found in Mac7 plants.
RNA extraction and real-time quantitative PCR analysis
Young leaves were collected from net house and glasshouse. Total RNA was extracted through the Trizol method60, and treated with RNase-free DNase (Promega, USA). The quality was assessed by gel electrophoresis. A 12μL sample with 100 ng/μL concentration, converted into cDNA using RevertAid Hminus First Strand cDNA Synthesis Kit (Thermo Scientific).
Based on the results of biotic stress expression profiling and host–pathogen protein interaction of GhCRKs, a set of genes was selected for qPCR analysis, and gene-specific primers were designed. Real-time PCR was performed using a Bio-Rad iCycler Thermal Cycler iQ5 and DNA Master SYBR Green I kit (Roche, Basel, Switzerland). Reactions were carried out in triplicate and each replicate consisted of 2 μL of cDNA (with concentration of100ng/μL), 0.5μL of each primer (concentration 10 μM/μL) and 5 μL SYBR Green Master Mix, making a final volume of 12 μL reaction. The PCR reactions were carried out using the following conditions: the initial temperature at 95 °C for 5 min, followed by 35 cycles of 95 °C for the 30 s, 58 °C for 30 s, and 72 °C for 1 min. Each biological sample was used in triplicates and the average expression value was calculated. The data ware normalized with the largest value of each panel making the highest relative expression as one.
Results
Gene organization of CRKs in diploid and tetraploid cotton species
The genome-wide analysis identified a total of 60, 74, 65, 120, and 118 CRK genes in Gar, Gra, Gher, Gbar, and Ghir, respectively (Tables S1–S5). The protein features were presented for each species, including protein length, molecular weight, charge, isoelectric point, and grand average of hydropathy. A summary of these features showed that the longest proteins sequence comprises 884 amino acids, while the shortest was composed of only 127 amino acid residues. Likewise, the molecular weight range was observed between 97.675 and 14.55 kDa, whereas the protein charge ranges between + 23.5 and − 15. Moreover, the isoelectric point ranged from 9.229 to 4.426, while the grand average of hydropathy range was from 0.172 to − 0.347 (Fig. S1).The chromosomal location and frequency of genes among the A-genome, D-genome, A-like genome, and D-like genome also demonstrated nearly similar gene density on respective chromosomes. For instance, the maximum number of genes was localized on Chr6, Chr10, Chr11 in the D-genome (Gra), and A-genome (A1; Gher, A2; Gar). Similarly, ChrA06 (Gar; 14 genes, Gher; 17, Ghir; 15 genes and Gbar with 14 genes), ChrD06 (Gra; 12 genes, Ghir; 14 genes, and Gbar; 15 genes), ChrD09, and ChrD10 (Gra; 11, Ghir; 14, and Gbar; 12 genes) possessed the highest number of genes in respective species. Additionally, most of the genes were found in clusters and were localized on the terminal arms of chromosomes. The gene clusters were randomly distributed along centromeres and telomeres (Fig. 1, Table S6). Overall, we found that most of the genes were localized on Chr6, Chr10, and Chr11 in all five species, representing their common locus the genomes.
Domain architecture, classification, and phylogenetic analysis of cotton CRKs
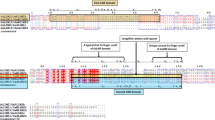
To provide a comprehensive study, we have introduced two new classification methods in cotton CRKs. The first was based on the type of domains presence and absence. In this classification system, all identified CRK genes were divided into 11 major classes. Through this classification, we have identified many Stress-antifungal (DUF 26)-associated functional domains such as ALMT, FUSC_2, Cript, DUF3403, FYVE, TauE, and Pkinase-tyr. Of these 11 classes, class VI (Stress-antifungal—Pkinase-tyr) has the largest number of genes, followed by class IV and VII. Class VI was the most commonly found class in all species with 73, 69, 36, 28, and 37 genes in Ghir, Gbar, Gar, Gher, and Gra, respectively. In contrast to shared classes, several species-specific classes (e.g. class_II only found in Gher, class_II only in Gra, class_IX only in Ghir, and class_X in Gbar) were also observed (Fig. 2, Table 1, Table S7).
The second classification method included the number of duplicated domains in addition to domain presence and absence. These classes were named as a subclass of CRK genes in Gossypium sp. All 437 genes were distributed into 43 sub-classes (I- XXXIII classes). The highest number of genes were observed in subclass IX (162 genes) with domain architecture Stress-antifungal__Stress-antifungal__DUF3403 and VII subclass (116 genes) with domains; Stress-antifungal__Stress-antifungal. These two classes were commonly found in all five species with the highest number of genes compared to other subclasses. In contrast to common domains architectures, unique and species-specific domain architectures were also observed (Fig. S2, Tables S8–S9).
The accumulative phylogenetic tree of all five species is divided into several major and minor clades. However, we did not observe any species-specific clade showing species diversity in the CRKs among Gossypium sp. The whole phylogenetic tree was divided into 51 subclades/clusters (clus), and of these, clus-XLVII had maximum genes with 15 CRKs, followed by clus-V, clus-V, clus-XXIII, and clus-VII with 14 CRKs. Similarly, clus-I, clus-XIV, and clus-XLIII consist of 13 CRKs and so on. Most of the clades possessed a range of 7 to 14 CRKs. The number of clades was related to subclasses of cotton CRKs (Fig. S3). Taken together, these CRK classifications identified several novel classes of species-specific and common members.
Evolutionary study of cotton CRKs with land plants
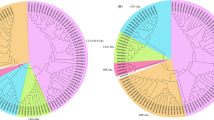
For the evolutionary study of Gossypium sp. with land plants, we included 2,026 CRKs from 35 plants, including mosses, bryophyte, gymnosperm, and angiosperm15 (Table S10). Of these land plants, Chlamydomonas reinhardtii, Coccomyxa subellipsoidea, Micromonas pusilla, Ostreococcus lucimarinus, Volvox carteri did not possess any CRK encoding genes. The identified CRKs were screened through Pfam and revised the steps mentioned for cotton classification. The conserved domain-pattern-based classification identified a total of 19 different patterns, thus classified into 19 subclasses (Table 2). Of these nineteen classes, class IV (ddCRRSP) and class X (ddCRK) were more common in all higher plants, while lower plants e.g., Marchantia polymorpha (liverwort) had only class I (sdCRRSP) followed by Selaginella moellendorffii (lycophyte) that possessed class I, class VIII (sdCRK) and class X (ddCRK). The Gossypium sp. showed several genus-specific classes including sdCRRSPdS, ddCRRSPD, tdCRRSP, sdCRKD, ddCRKF, ddCRKS and qdCRKD. These classes are only found in cotton species showing their diversity with other dicot plants (Fig. 3, Table S11).
The comparative genomics summarized that all CRKs from 5 species of cotton plants were divided into 52 orthogroups covering 416 genes (95.2% of genes in orthogroups) with only 21 unassigned genes (4.8% of genes). Of these, 40 orthogroups were shared by all five species, while none of the orthogroups were species-specific. Overall, the mean and medians were recorded as 8 and 7 orthogroups, respectively. At the species level, however, we observed that G. arboreum shared more orthologs with G. barbadense (92 orthologs) and G. hirsutum (89 orthologs) as compared to G. raimondii (58 orthologs) and G. herbaceum (57 orthologs). Similarly, G. barbadense shared a higher number of orthologs with G. hirsutum (100 orthologs), followed by G. raimondii (66 orthologs), G. arboreum (56 orthologs), and G. herbaceum (54 orthologs) (Fig. 4A–C).
Summary of OrthoFinder analysis of CRK gene family. (A) The species of Gossypium sp. (B) Percentage of genes in orthogroups. (C) The number of species-specific orthogroups. (D) Heat map showing shared orthogroups. (E) Orthologs multiplicity. (F) Gene duplication events per species. (G) Gene duplication per internal and terminal nods of the species-based-phylogenetic tree. (H) Circos plot of Gar, Gher, Gra with Gbar. (I) Circos plot of Ghir with Gar, Gher and Gra. Tca; T. cacao.
The species-wise orthogroups distribution represented that most of the identified genes belonged to one of 52 orthogroups, i.e., G. arboreum (84.6% of total GaCRKs), G. raimondii (88.5% of total GrCRKs),) and G. herbaceum (86.5% of total GheCRKs), G. barbadense (84% of total GbCRKs), and G. hirsutum (96.2% of total GhCRKs). The species-wise relationship demonstrated remarkable relation among five species. We also drew one-to-one, one-to-many, many-to-one, and many-to-many species relationships. We concluded that a small number of genes contributed in one-to-one and many to one, while a higher number of genes showed one-to-many and many-to-many with a concluding close relationship of five species (Fig. 4D,E).
The species-wise phylogenetic tree with Theobroma cacao (T. cacao) as an outgroup, demonstrated that G. arboreum (A2-genome) has close relation with G. hirsutum, followed by Gra (D-genome). The gene duplication event was also predicted at all internal and terminal nodes. N0 node represented the common ancestor of all cotton species with T. cacao, demonstrated 69 duplications with 100% confidence and 40 duplications with 50% confidence. Emerging from the N0 node, T. cacao gained 16 duplications, Gbar gained 22, Gra gained 12 and Ghir gained 24 duplication events. While the Gher (A1-genome) and Gar (A2 genome) did not show any duplication events. (Fig. 4F,G). The chromosomal location collinearity suggested that the Ghir has more syntenic blocks within its subgenome (A and D genomes) followed by Gar (A-genome) while the G. barbadence showed more collinearity lines with Gher (A1-genome) and Gra (D-genome) (Fig. 4H,I).
De-novo motif discovery and functional sites prediction and tissue-specific expression profiling in diverse cotton species
The de-novo MEME motif analysis identified a total of 15 conserved motifs in all-cotton CRK genes (Fig. S4). Of these, motif_13, motif_9, motif_2, and motif_12 are highly conserved in all CKRs. However, motif_1, motif_4, motif_11, and motif_6 were gene-specific. In addition, the functional motif sites prediction through PROSITE identified, a total of 26 important functional motifs including asn_glycosylation, myristyl, ck2_phospho, rgd, pkc_phospho, protein_kinase_dom, protein_kinase_atp, camp_phospho, protein_kinase_st, amidation, leucine_zipper, hma_1, phe_rich, atp_gtp_a, ser_rich, n6_mtase, pro_rich, tonb_dependent_rec_1, peroxidase_1, prokar_lipoprotein, zf_fyve, microbodies_cter and prenylation. Overall, we concluded that the different groups of CRKs possessed different functional and structural motifs that might provide functional diversity (Table S12).
The RNA-seq data analysis of CRK genes in G. arboreum showed distinctive expression patterns in diverse tissues (leaf, stem, and root) at different time intervals (10 DPA, 15 DPA, and 20 DPA). Some genes, including GaCRK02, GaCRK09, GaCRK52, and GaCRK29, showed increased transcript levels in the leaf than stem or root. Similarly, several genes (GaCRK02, GaCRK24, GaCRK03, and GaCRK38) had elevated transcript levels in the stem, while others exhibited higher mRNA levels in the root (GaCRK24, GaCRK02, GaCRK43, and GaCRK07). In addition, the expression profiling in the ovule at 10 DPA, 15 DPA, and 20 DPA demonstrated remarkable differences in the expression of CRK genes. For instance, in ovule development at 15 DPA, GaCRK03, GaCRK02, and GaCRK38 showed the highest expression levels, and this set of genes also showed similar results at 15 DPA and 20 DPA in ovule and fiber development (Fig. S5A, Table S13). In summary, most of the genes demonstrated tissue-specific expression, However, GaCRK02 showed high transcripts in all tissues at different time intervals.
The G. raimondii RNA-seq analysis of CRKs in different tissues such as seed, fiber, ovule, and leaf at different time intervals (10 DPA, 20 DPA, 30 DPA, and 40 DPA) was also presented. Results demonstrated that GrCRK11, GrCRK67, and GrCRK52 showed higher transcript levels in seed germination at 10 DPA in comparison with 20 DPA, 30 DPA, and 40 DPA data. Similarly, some genes (GrCRK67, GrCRK21, GrCRK24, and GrCRK42) exhibited increased transcript levels at 20 DPA, while others showed higher mRNA levels at 30 DPA (GrCRK21, GrCRK67, GrCRK24, and GrCRK42) and 40 DPA (GrCRK63, GrCRK21, GrCRK11, and GrCRK67). Similar expression patterns were also observed in fiber development at 10 DPA (GrCRK24, GrCRK11, GrCRK46, and GrCRK18) and 20 DPA (GrCRK06, GrCRK21, GrCRK67, and GrCRK24) (Fig. S5B, Table S14). Like the G. arboreum CRKs expressions, the G. raimondii CRKs also demonstrated clusters of genes co-expressing in different tissues. However, GrCRK67 showed its putative role in all tissues.
Gossypium hirsutum is known as upland cotton, and these species produce more than 90% of the world's raw cotton. Therefore, several transcriptomics data are available for this plant. The tissue-specific RNA-seq data included different tissues (leaf, bract, sepal, stem, root, ovule, torus, filament, petal, and anther). The RNA-seq results demonstrated diverse tissue-specific expression patterns of CRKs in various tissues. Some prominent genes (GhCRK044, GhCRK110, GhCRK068, GhCRK086, GhCRK107, and GhCRK084) exhibited high transcript values in filament, petal, and anther. However, a few genes associated with similar clusters depicted higher expression in stem (GhCRK77, GhCRK053, GhCRK013, GhCRK015, and GhCRK018), ovule (GhCRK007, GhCRK013, GhCRK015, GhCRK014, and GhCRK005), torus (GhCRK018, GhCRK015, GhCRK005, and GhCRK044) and in bract (GhCRK094, GhCRK053, GhCRK060, GhCRK077, GhCRK110, and GhCRK078) tissue. These differential expressions indicate the role of CRK genes in the growth and development of multiple tissues (Fig. 5A, Tables S15–S16).
The RNA-seq data of CRK genes under different abiotic stresses including salt, drought, heat, and cold stresses was demonstrated at different time intervals (1 h, 3 h, 6 h, and 24 h) and days intervals (0 DPA, 1 DPA, 3 DPA, 10 DPA, 15 DPA, and 20 DPA) (Tables S17–S21). The comparative expression profiling revealed several differentially expressed gene clusters displaying increased transcripts values at corresponding stress conditions. For instance, a cluster of genes including GhCRK060, GhCRK053, GhCRK083, and GhCRK110 showed high expression under all stresses including salt, drought, heat, and cold. However, most of the CRKs depicted tissue-specific expressions. For instance, GhCRK109 was highly expressed under salt stress while did not show induced in other stresses. Similarly, the GhCRK053 gene is highly induced under drought, cold and heat stresses rather than salt stress. The clusters of genes that showed their co-expressions and co-occurrences under specific stresses might have an accumulative role in cotton plant adaptation during environmental stresses (Fig. 5B).
Protein–protein interaction network and host–pathogen interaction
The protein–protein interaction network of GhCRKs provided endogenous protein interactions including, experimentally determined interactions, gene fusion, gene co-occurrence, co-expression, and protein homology. Of 118 GhCRKs, only a few proteins showed internal interactions. For instance, GhCRK067 has the highest number of interactions including GhCRK067-GhCRK025 and GhCRK67-GhCRK048 possessing experimentally validated interactions, GhCRK067-GhCRK028 complex has three types of correlation i.e. Co-expression, protein homology, and text mining. Similar interactions were also observed in the GhCRK084-GhCRK028 complex (Fig. S6). We selected ten genes for further host–pathogen interaction analysis based on the differential expression of CRKs in Mac7 and NIAB-Karishma under CLCuD stress. The protein–protein interaction probability analysis demonstrated strong interaction of most of the up-regulated genes in Mac7 to the Begomovirus protein, including GhCRK082 (strongly interaction probability with AC1, AC2, AC3, AC4, AV2, and C5) and GhCRK087 (strongly interaction probability with AC1, AC2, AC3, and AV2) had a probability value of more than 0.9. In comparison, almost all other genes had greater than 0.5 values, which is significant for protein–protein interaction (Fig. S7, Table S22). Furthermore, the sequence-based interactions of GhCRKs with viral proteins were also demonstrated with host–pathogen protein–protein interaction with ZDOCK molecular docking. The 3D host–pathogen protein docking analysis demonstrated the interaction network between host and pathogen amino acidic residues. The upregulated genes and their interaction with CLCuD viral proteins confirmed their direct interaction. The GhCRK059 and AC2 interaction provided the active residues and their bonding types. As such, the GhCRK059 protein (A chain) with residues Leu23, Arg22, Ser24, and His25 interacted with AC2 protein (Q chain) at Cys36, Asp110, and Ser39 through hydrogen bond (green lines) and salt bridge (red lines). Similar results were observed in GhCRK087-AC3, GhCRK087-AV2, GhCRK082-AC2, GhCRK082-AV2, and GhCRK082-AC3 complexes. However, the number of bonds and types of bonds varied from complex to complex e.g., the highest number of interactions was found in GhCRK082-AC3, followed by GhCRK082-AC2 (Fig. 6, Fig. S8).
Molecular docking of host–pathogen interaction. (A) GhCRK21-AC2 complex, (B) GhCRK21-AC2 complex 2D interaction graph, (C) GhCRK45-AC3 complex, (D) 2D interaction graph of GhCRK45-AC3 complex,(E) GhCRK45—V2 complex, (F) 2D interaction graph of GhCRK45—AV2. The green color represents viral proteins and blue is for Host CRKs. Green lines; hydrogen bond, red lines; salt bridge.
SNPs and InDels variants in CRKs of resistant G. hirsutum
Mac7 is a tolerant G. hirsutum accession, which is developed by USDA by breeding program. To find genetic variation and transcriptomics variation, we have identified SNPs and InDels associated with CRKs in Mac7 and Coker 312. The genome-wide-genetic variation in Mac7 identified a total of 192 and 208 genes having SNPs and InDels concerning TM-1 reference G. hirsutum genome, respectively. Similarly, a total of 62 and 192 genes with SNPs and InDels were found in Coker 312, respectively. The comparative study of Mac7 and Coker 312 identified several unique SNPs and InDels in different genomics regions with different effects. For instance, 82 nonsynonymous, 64 synonymous, and 51 in 3’ UTR, SNPs were observed Mac7 (Table S23). In addition to genomic region-based variants, we also categorized variants into impact-based levels e.g. high, low, moderate, and modifier. In the case of variants by impacts, a total of 12 and 23 genes showed high impact SNPs and InDels in Mac7, respectively. While only 3 SNPs and 17 InDels associated genes were found in Coker 312 under the same variant impact level (Table S24). In summary, we have observed many differences in CRK gene sequences of the Mac7 and Coker 312 (Fig. S9). There were several unique SNPs and InDels in CRK genes of Mac7 accessions which could be the source of resistance to different biotic and abiotic stresses.
Micro-RNA and their target sites prediction in CRK genes
The miRNA target site prediction analysis demonstrated that most of the CRK genes possess miRNA target sites. However, the five species understudy showed somehow common and unique miRNA families. To provide deep analysis, the CRK gene-targeted miRNA, all identified miRNA target sites were categorized into family-based and family-member-based in all five species. A total of 30, 2, 1, 117, and 30 miRNA families were detected in CRK genes of G. arboreum (Gar), G. raimondii (Gra)61, G. herbaceum (Gher), G. hirsutum (Ghir), and G. barbadense (Gbar)62, receptively. In these miRNA families, a total of 83, 3, 1, 346, and 150 miRNAs were detected in Gar, Gra, Gher, Ghir, and Gbar, respectively. In Gar, some miRNA families, including Gar-miR172 (59 target sites, Gar-miR396 (22 target sites), and Gar-miR1373 (14 target sites), have a high number of target sites in GaCRK genes. Of the Gar-miR172 family, Gar-miR172c (7 target sites), Gar-miR172d (7 target sites), and Gar-miR172e (7 target sites) had a higher number of target sites (Fig. S10A, Table S25). In the G. raimondii CRK gene, only two miRNA families were detected, including Gra-miR172 (16 miRNA target sites) and Gra-miR482 (3 miRNA target sites). (Fig. S10B, Table S26). In G. herbaceum, only a single miRNA was detected (Fig. S10C, Table S27). Since the number of CRKs is higher in tetraploid cotton, we found an increased number of miRNAs. A total of 10 miRNA families were predicted in G. hirsutum with the highest number of target sites by Ghi-miR172 (296 miRNA target sites), Ghi-miR394 (60 miRNA target sites), and Ghi-miR1404 (58 miRNA target sites), and so on. At the family member level in the Ghi-miR172 family, three major targeting members were observed as Ghi-miR172d (22 miRNA target sites), Ghi-miR172e (22 miRNA target sites), and Ghi-miR172f (22 miRNA target sites) (Fig. S10D, Table S28). Similarly, G. barbadense, Gba-miR172, Gba-miR156, and Gba-miR395 families had the highest number of miRNA targets sites within CRK genes (Fig. S10E, Table S29). The comparative study of miRNA families and members among five species demonstrated that both common and species-specific miRNAs. The most common miRNA family in CRK genes was miR172 and miR156 among all species. However, we also observed some species-specific miRNA families. For instance, Ghir had 85 unique miRNA families, Gar had seven unique miRNA families, and Gbar had only seven (Fig. S11A,B). Similar findings were also observed in miRNA family members among the five species. Overall, cotton CRK genes possessed more miRNA target sites for the miR172 family that might be the main functional regulator of cotton CRK genes.
Expression profiling of identified miRNA families
The expression profiling of miRNA families and their members provides significant information about the CRK genes regulation. Thus, we have identified the expression level of miRNA families in different tissues of G. arboreum, G. barbadense, and G. hirsutum.
For the expression profiling of G. arboreum miRNA, we included fiber, flower, and leaf tissues. In fiber tissue, the highest expression of Gar-miR172, Gar-miR164, and Gar-miR3476 was observed. Similarly, Gar-miR172, Gar-miR535, and Gar-miR164 showed elevated expression in flower, while Gar-miR156, Gar-miR172, and Gar-miR535 displays increased expression in leaf (Fig. 7A, Table S30). In G. barbadense, we only found data for two tissues, i.e., fiber and apical shoot. The highest expression of miRNA (Gba-miR172, Gba-miR164, and Gba-miR166) was observed in fiber, whereas Gba-miR156, Gba-miR166, and Gba-miR172 were found to be elevated in the shoot apical (Fig. 7B, (Table S31). In summary, we discovered a strong role of miR172, miR156, and miR159 in regulating CRKs in G. barbadense plants. The expression profiling of G. hirsutum miRNA included different tissues, and most of the miRNAs showed tissue-specific expression. For instance, highest expression of miRNAs was observed in anther (Ghi-miR166, Ghi-miR172, and Ghi-miR156), embryogenic (Ghi-miR156, Ghi-miR166, and Ghi-miR164), fiber (Ghi-miR166, Ghi-miR164, and Ghi-miR3476), hypocotyls (Ghi-miR156, Ghi-miR166, and Ghi-miR3476), leaf (Ghi-miR166, Ghi-miR1441, Ghi-miR159, and Ghi-miR156), ovule (Ghi-miR166, Ghi-miR1441, and Ghi-miR159), root (Ghi-miR156, Ghi-miR166, and Ghi-miR1383), apical shoot (Ghi-miR156) and in apexes stem (Ghi-miR159, Ghi-miR166, and Ghi-miR319) (Fig. 7C, Table S32). In conclusion, we have observed that miR172 has high expression in most of the tissues like fiber, flower, and apical shoots, displaying its role in the regulation of the CRK gene.
Expression profiling and RT-PCR analysis of GhCRKs under CLCuD in resistant and susceptible G. hirsutum
We used RNA-seq experimental data from Mac7 (G. hirsutum accession, resistant to CLCuD)50 and NIAB-Karishma (a mutant Coker 312 G. hirsutum, highly susceptible to CLCuD)49 for expression analysis (Fig. S12). The RNA-seq data include viruliferous whitefly infestation for cotton leaf curl virus disease (CLCuD), Pakistan's threat to cotton production. The expression profiling and comparison demonstrated that 86 CRK genes and 32 genes are expressed in Mac7 and NIAB-Karishma, respectively. The comparative study revealed that most CRK genes showed increased expression in resistant (Mac7) than susceptible NIAB-Karishma. For instance, GhCRK026, GhCRK013, GhCRK007, GhCRK116, GhCRK108, GhCRK099, GhCRK082, GhCRK072, and GhCRK096 were differentially upregulated in resistance under CLCuD disease treatment (Fig. S13, Table S33). The quantitative real-time expression analysis of selected genes displayed intriguing findings. Of the nine selected genes (Table S34), only three genes (GhCRK093, GhCRK82, and GhCRK096) showed some level of expression in Coker 312 (susceptible accession) and their transcripts level decreased when treated with CLCuD. However, all 9 genes showed increased expression in Mac7 (resistant accession) in control as well as infested sample, and some genes (GhCRK059, GhCRK081, and GhCRK087) were upregulated under CLCuD (Fig. 8). Generally, we concluded that the CRK genes in Mac7 and Coker are shown to be involved in CLCuD stress.
Relative expressions of GhCRKs under cotton leaf curl disease. Mac7_C; resistant control, Mac7_WI; resistant infested with viruliferous whitefly, Coker 312_C; susceptible control, Coker 312_WI; susceptible infested with viruliferous whitefly. Error bars represent the SD of three independent experiments.
Discussion
Cotton (Gossypium sp.) is a worldwide economical crop that produces raw fiber and seed oil for the textile and oil industries. But the environmental stresses including biotic (insect, pest, virus, and bacteria) and abiotic (drought, salinity, heat, and cold) are limiting its growth and yield. Thus, the improvement of resistant genetic makeup is essential for high-quality cotton production. CLCuD is one of the major biotic stresses in Asia such as Pakistan and India and this deadly virus decreases cotton yield several-fold every year. Therefore, identification, characterization, and functional analysis of stress-responsive genes are the top targets of cotton researchers. Several genome-wide association studies have been conducted to find important genes involved in different agronomical traits such as fiber yield and improvement, gossypol content, drought, and salt stress-resistant. Similarly, many important resistant QTLs and markers against different abiotic and biotic stresses have been discovered63. However, very little is known about the CLCuD resistant mechanism. The current study also provided important data for further functional analysis against CLCuD.
The biotic stress signal in plants observed by pattern recognition receptors (PRRs) also plays a vital role in activating plants' immunity. PAMPs are the biotic stress signals that activate a combination of immune receptors complexes and plant immune response signaling pathways64. The pathways that respond to early immune signaling are kinase and transcriptional gene regulation65. Antifungal proteins are also called CRKs (Cysteine (C)-rich receptor-like kinases gene) and DUF26 or Gnk-2. CRKs are an important class of receptor-like kinase (RLKs) that play vital roles in disease resistance in plants. Despite the known role of CRKs in plant resistance17,21,22,66,67,68, there is a big gap in a genome-wide comparative study of CRKs in Gossypium species, which was covered in this study. The genomic analysis effectively transfers knowledge from one taxon to another, allowing for a faster pace of gene discoveries associated with disease resistance.
The current study identified CRKs in five Gossypium sp. and classified them according to domains architectures. A total of 437 CRK genes have stress-antifungal domains in G. arboreum (60 genes), G. raimondii (74 genes), G. herbaceum (65 genes), G. hirsutum (118 genes), and G. barbadense (120 genes). A similar study of CRK was also conducted by Ting et al. and Hussain et al.37,69, but several gaps were filled by this study as we identified an increased number of CRK genes compared to the previous studies. In addition, several additional bioinformatics and expression analyses were also added in the current work. The number of CRKs in tetraploids was not more than two-fold of diploid species that may be due to gain and loss of the gene during polyploidization of two diploid genomes (A and D genome) to make the tetraploid genomes (AAtDDt)70. Furthermore, the segmented and tandem duplications influence the development of multiple CRKs gene families, including the RLKs family71. The gene structure analysis identified exon, intron, and UTR regions of the genes. The length of intron and exon is related to the phylogenetic tree's construction, and it is particularly high in G. raimondii72. The comparative analysis showed both differences and similarities in the exon number that might be related to their function and conservation73. Chromosomal mapping of CRKs showed their abundance on a few chromosomes like Chr6, Chr10, and Chr11 in all five species. Similar chromosomal mapping was also reported by Ting et al.69. The region behind the higher density of genes on some species might be due to segmental duplications that occurred between non-allelic chromosomes in cotton. Furthermore, the DUF26 containing genes normally show tandem duplication74.
In this study, we have classified CRKs in all five species in three different ways, conserved domains absence and presence (DAP), conserved domain repeats (DR), 3rd classification based on Aleksia et al.15. Based on the first and second classifications, we have identified several DUF26 associated decoy domains, like ALMT (aluminum-activated malate transporter domain)75, FusC_2 (Fusaric acid resistance protein-like domain)76, Cript (Microtubule-associated protein domain), FYVE (evolutionarily conserved double-zinc-binding domain) which are functionally well-characterized protein domains and are involved in diver biological function including efflux of organic acids77, salt stress tolerance and the regulation of malic acid content78, linkage of mRNA transporter to endosome trafficking79. Such resistance decoy domains also reported in Gossypium37 may provide additional features to CRK genes for plants adaptation. Molecular phylogenetic analysis and OrthoFinder results suggested a significantly divergent evolutionary history of CRK genes in five species. The species-based phylogenetic tree of CRKs suggested G. arboreum as the ancestor of other Gossypium sp. and the othogroups also found some species-specific groups. However, the five species showed a close relationship in sequence similarity, possibly due to the origination of CRKs from common ancestors. The evolution of genes is mediated by sequence exchange, tandem or segmental duplication events, or gene conversion80,81,82.
A single-nucleotide polymorphism (SNPs) is the simplest form of genetic variation among individuals that can prompt minor changes in phenotypic, physiological, and biochemical characteristics. These mutations in the gene sequence alter the amino acid sequence, which may change the function of the gene. Several SNPs were identified and used as a genetic marker for the identification of qualitative trait loci (QTLs) associated with multiple agronomic features of cotton including fiber quality and quantity, resistance to biotic and abiotic stresses83,84,85,86,87. However, very little is known about SNPs associated with biotic stress resistance. So, we also have found several SNPs in CRK genes of resistant accession of G. hirsutum in comparison with the Coker 312 (highly susceptible) and TM-1 reference genome. We suggest that these SNPs may have a significant role in plant adaptation under CLCuD. However, further experimental validation is required to confirm their role as selection markers.
miRNA contains 17 to 24 nucleotides (nt) and is an important gene regulatory factor in plants88. miRNAs take part in diverse biological functions of plants at different transcriptional and translational levels88,89,90,91. They also play essential roles in developing immunity against pathogens succeeding the endogenous defense-related genes and down-regulating the pathogens of the exogenous viral plants92,93,94. The miRNA target sites in CRKs provide essential primary data for understanding CRKs regulation under various stresses. In cotton CRKs, we found different miRNA families in diverse cotton species such as miR172, miR1373, miR169, and miR164 in GaCRKs, whereas miR172, miR159, miR169, and miR397 in GhCRKs. The genotype-depended response of miRNA to biotic and abiotic stresses varies from cultivar to cultivar95. Another study of miRNA microarray in cotton found high expression of miR156, miR169, miR535, and miR827 under salinity treatment96. The miR172b-SSR was used as a biomarker for identifying the different responses of rice cultivars under salt stress97. In agreement with the literature, several miRNA families were identified which could target CLCuDV genes with perfect and near-perfect complementarity98. In addition, the miR172 initiates floral growth and modifies reproductive growth and the co-regulation of miR156 and miR172 in the origination and improvement of cotton plants99. So, the identification of miRNAs in cotton CRKs would be helpful for understanding the post-transcriptional regulation of CRKs under diverse stress conditions. We also summarize based on the miRNA target site prediction and expression profiling of miRNA, there might a strong correlation between miRNA expression and functional regulation of cotton CRK genes. As we have observed that CRKs have the highest target side for the miR172 family and in the meanwhile the miR172 family has high expression in most of the tissue, demonstrating its role in the regulation of various biological mechanisms.
The difference in the number of genes and classes among 35 plants species is due to the expansion of CRK families through small-scale duplication, genome fractionation, and genetic drift which cause due to whole genome multiplications16. One prediction suggests that the duplicated genes under dosage balance exhibit fewer expressions than other duplicates and RLKs can function not only in defense but also in development and abiotic stress responses100,101. Hence, to determine the functional conservation and putative role of CRKs in cotton development and adoption under stresses, publicly available expression data of tissues and stress treatments were used and highlighted several tissue-specific and stress-specific expressions of CRKs in cotton. For instance, a cluster of CRKs; GhCRK053, GhCRK083, GhCRK094, GhCRK038, GhCRK110, GhCRK039, GhCRK041, GhCRK068, GhCRK093, and GhCRK013, induced transcriptionally under salt, drought, heat, and cold stresses in G. hirsutum. Previous studies reported that AtCRKs are transcriptionally induced under abiotic stresses such as salt, drought, UV light, heat, salicylic acid17,18,19,20,21. In agreement with such previous studies of CRKs in Arabidopsis, we also found important several stress-responsive cotton CRKs. In addition to abiotic stresses, we also reported the important cotton CRKs response under biotic stresses including cotton leaf curl disease for instance the G. hirsutum CRKs; GhCRK026, GhCRK013, GhCRK007, GhCRK116, GhCRK108, GhCRK099, GhCRK082, and GhCRK072 differentially upregulated in resistant accession while down-regulated in susceptible accession under CLCuD disease treatment. The genetic variants analysis also identified several SNPs and InDels in these putative genes. Furthermore, the quantitative real-time expression analysis also validated the RNA-seq analysis of GhCRKs. All selected GhCRKs were transcriptionally upregulated in resistant accession while either down-regulated or did not show any transcript in the susceptible accession. In literature, it is reported that a subset of CRKs is strongly induced in response to pathogens and PAMPs treatments19,20 and overexpression of AtCRK4, AtCRK5, AtCRK6, AtCRK13, and AtCRK36 showed enhanced resistance to the bacterial pathogen Pseudomonas syringae as well as also activated the early and late PTI responses17,21,22. Henceforth, the comparative expression of CRKs in resistant and highly susceptible cotton provided important CRKs candidates. The coordination of these CRKs during plant immune response suggested that they cooperate in plant defense signaling. Furthermore, the molecular docking of CRKs with CLCuD viral proteins also demonstrated their direct interaction. As CRK proteins possess extracellular domains, which are involved in protein–protein interaction and signal perceptions12, the transmembrane domains and intracellular domains transduce and activate MAPK pathways for activation of the plant. The host–pathogen interaction and expression data showed coordination of these putative genes in plant immune signaling. Thus, the GhCRK057, GhCRK059, GhCRK058, GhCRK081, GhCRK008, and GhCKR087 might be a potential marker for CLCuD resistant genotype. The current study provided a deep insight into CRKs in Gossypium sp. The different ploidy level of Gossypium species has different resistance level, for instance, the diploid species like G. arboreum, G. raimondii, and G. herbaceum are naturally resistant to several biotic and abiotic stresses while the tetraploid cotton-like G. hirsutum is susceptible to multiple stresses and the G. barbadense is highly susceptible to environmental stresses. Thus, the comparative study of stress-responsive genes CRKs in cotton is essential for improving cotton growth and development.
Conclusion
The current study identified a total of 437 Cysteine-rich receptor-like kinases (CRKs) encoding genes in five Gossypium sp. The structural and domain-based classification identified several novel domain architectures in Gossypium sp. The genome mapping and genetic diversity (SNPs and InDels) provided important data for cotton breeders and the expression profiling under different environmental stresses and their validation through qPCR under CLCuD demonstrated a putative role in cotton growth and development. The miRNA target site prediction will help to understand the regulation of CRKs in specific tissues. We have provided detailed computational and experimental studies on CRKs in the five species; however, further individual gene functional analysis is required to understand the CRKs mechanism in cotton plants' adaptation.
Data availability
Source data for all the graphs included in this paper are available as Supplementary Data in Excel format. All other data are available from the corresponding author upon reasonable request. It is also stated that there are no ethical issues that required permissions or licenses to complete this work.
References
Netea, M. G. & van der Meer, J. W. M. Trained immunity: An ancient way of remembering. Cell Host Microbe 21, 297–300 (2017).
Ranf, S. Pattern recognition receptors: Versatile genetic tools for engineering broad-spectrum disease resistance in crops. Agronomy 8, 134 (2018).
Mishra, B., Kumar, N. & Mukhtar, M. S. Network biology to uncover functional and structural properties of the plant immune system. Curr. Opin. Plant Biol. 62, 102057 (2021).
Spears, B. J. et al. Direct regulation of the EFR-dependent immune response by Arabidopsis TCP transcription factors. Mol. Plant Microbe Interact. 32, 540–549 (2019).
Mishra, B., Sun, Y., Ahmed, H., Liu, X. & Mukhtar, M. S. Global temporal dynamic landscape of pathogen-mediated subversion of Arabidopsis innate immunity. Sci. Rep. 7, 1–13 (2017).
Yang, X., Deng, F. & Ramonell, K. M. Receptor-like kinases and receptor-like proteins: Keys to pathogen recognition and defense signaling in plant innate immunity. Front. Biol. 7, 155–166 (2012).
Zhu, S., Fu, Q., Xu, F., Zheng, H. & Yu, F. New paradigms in cell adaptation: decades of discoveries on the CrRLK1L receptor kinase signalling network. New Phytol. 232, 1168–1183 (2021).
Cristina, M. S., Petersen, M. & Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649 (2010).
Nürnberger, T. & Scheel, D. Signal transmission in the plant immune response. Trends Plant Sci. 6, 372–379 (2001).
Zhu, S., Fu, Q., Xu, F., Zheng, H. & Yu, F. New paradigms in cell adaptation: Decades of discoveries on the CrRLK1L receptor kinase signalling network. New Phytol. Phytol. 232, 1168–1183 (2021).
Ahmed, H. et al. Network biology discovers pathogen contact points in host protein-protein interactomes. Nat. Commun. 9, 1–13 (2018).
Smakowska-Luzan, E. et al. An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553, 342 (2018).
Vaid, N., Macovei, A. & Tuteja, N. Knights in action: Lectin receptor-like kinases in plant development and stress responses. Mol. Plant 6, 1405–1418 (2013).
Ye, Y. et al. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 36, 235–242 (2017).
Aleksia, V. et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2, 56 (2019).
Vaattovaara, A. et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2, 56 (2019).
Chen, K., Du, L. & Chen, Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53, 61–74 (2003).
Chen, K., Fan, B., Du, L. & Chen, Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 56, 271–283 (2004).
Wrzaczek, M. et al. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10, 1–19 (2010).
Bourdais, G. et al. Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 11, e1005373 (2015).
Yeh, Y.-H., Chang, Y.-H., Huang, P.-Y., Huang, J.-B. & Zimmerli, L. Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6, 322 (2015).
Acharya, B. R. et al. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50, 488–499 (2007).
Vaattovaara, A. et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2, 1–18 (2019).
Wang, K., Wendel, J. F. & Hua, J. Designations for individual genomes and chromosomes in Gossypium. J. Cotton Res. 1, 3 (2018).
Wang, K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103 (2012).
Wang, M. et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229 (2019).
Li, F. et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530 (2015).
Li, F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572 (2014).
Udall, J. A. et al. De novo genome sequence assemblies of Gossypium raimondii and Gossypium turneri. G3 9, 3079–3085 (2019).
Paterson, A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427 (2012).
Zhang, J. et al. Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 187, 147–160 (2012).
Zehr, U. B. Cotton: Biotechnological Advances Vol. 65, 256 (Springer Science & Business Media, 2010).
Yu, J. et al. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, D1229–D1236 (2014).
Hu, Y. et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748 (2019).
Yu, J. et al. CottonGen: The community database for cotton genomics, genetics, and breeding research. Plants 10, 2805 (2021).
Mistry, J. et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2020).
Hussain, A. et al. Genome-wide identification and classification of resistance genes predicted several decoy domains in Gossypium sp. Plant Gene 24, 100250 (2020).
Guo, A.-Y., Zhu, Q.-H., Chen, X. & Luo, J.-C. GSDS: A gene structure display server. Yi Chuan 29, 1023–1026 (2007).
Bailey, T. L., Johnson, J., Grant, C. E. & Noble, W. S. The MEME suite. Nucleic Acids Res. 43, W39–W49 (2015).
Hulo, N. et al. The PROSITE database. Nucleic Acids Res. 34, D227–D230 (2006).
Emms, D. M. & Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14 (2019).
Davim, J. P. Diamond tool performance in machining metal–matrix composites. J. Mater. Process. Technol. 128, 100–105 (2002).
Azad, A., Pavlopoulos, G. A., Ouzounis, C. A., Kyrpides, N. C. & Buluç, A. HipMCL: A high-performance parallel implementation of the Markov clustering algorithm for large-scale networks. Nucleic Acids Res. 46, e33–e33 (2018).
Kelly, S. & Maini, P. K. DendroBLAST: Approximate phylogenetic trees in the absence of multiple sequence alignments. PLoS ONE 8, e58537 (2013).
Lefort, V., Desper, R. & Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800 (2015).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2: Approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Zhu, T. et al. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 17, 1–9 (2017).
Naqvi, R. Z. et al. Transcriptomic analysis of cultivated cotton Gossypium hirsutum provides insights into host responses upon whitefly-mediated transmission of cotton leaf curl disease. PLoS ONE 14, e0210011 (2019).
Zaidi, S. S. E. A. et al. Molecular insight into cotton leaf curl geminivirus disease resistance in cultivated cotton (Gossypium hirsutum). Plant Biotechnol. J. 18, 691–706 (2020).
Basit, A. H., Abbasi, W. A., Asif, A. & Minhas, F. U. A. A. Training host-pathogen protein-protein interaction predictors. J. Bioinform. Comput. Biol. 16, 1850014 (2018).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 9, 1–8 (2008).
Pierce, B. G. et al. ZDOCK server: Interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773 (2014).
Biovia, D. S. BIOVIA discovery studio visualizer. Softw. Version 20, 779 (2016).
Laskowski, R. A. et al. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2010).
Zhao, T. et al. Genome-wide analysis of genetic variations between dominant and recessive NILs of glanded and glandless cottons. Sci. Rep. 9, 1–10 (2019).
Yi, X., Zhang, Z., Ling, Y., Xu, W. & Su, Z. PNRD: A plant non-coding RNA database. Nucleic Acids Res. 43, D982–D989 (2015).
Guo, Z. et al. PmiREN: A comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 48, D1114–D1121 (2020).
Dai, X., Zhuang, Z. & Zhao, P. X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res 46, W49–W54 (2018).
Rio, D. C., Ares, M., Hannon, G. J. & Nilsen, T. W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harbor Protoc. 2010, 5439 (2010).
Karasev, A. V. & Gray, S. M. Continuous and emerging challenges of Potato virus Y in potato. Annu. Rev. Phytopathol. 51, 571–586 (2013).
Cho, H. & Winans, S. C. VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc. Natl. Acad. Sci. USA 102, 14843–14848 (2005).
Majeed, S. et al. Role of SNPs in determining QTLs for major traits in cotton. J. Cotton Res. 2, 5 (2019).
Macho, A. P. & Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272 (2014).
Boller, T. & Felix, G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 (2009).
Ederli, L. et al. The Arabidopsis thaliana cysteine-rich receptor-like kinase CRK20 modulates host responses to Pseudomonas syringae pv. tomato DC3000 infection. J. Plant Physiol. 168, 1784–1794 (2011).
Zhang, X. et al. Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae. Plant Physiol. Biochem. 73, 383–391 (2013).
Yadeta, K. A. et al. A cysteine-rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiol. 173, 771–787 (2017).
Li, T.-G. et al. Genome-wide identification and functional analyses of the CRK gene family in cotton reveals GbCRK18 confers verticillium wilt resistance in Gossypium barbadense. Front. Plant Sci. 9, 1266 (2018).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010).
Liu, J. et al. Soybean kinome: functional classification and gene expression patterns. J. Exp. Bot. 66, 1919–1934 (2015).
He, D. et al. Identification and analysis of the TIFY gene family in Gossypium raimondii. Genet. Mol. Res. 14, 10119–10138 (2015).
Liu, Y. et al. Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol. Biochem. 64, 92–98 (2013).
Lehti-Shiu, M. D., Zou, C., Hanada, K. & Shiu, S.-H. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26 (2009).
Liu, J. & Zhou, M. The ALMT gene family performs multiple functions in plants. Agronomy 8, 20 (2018).
Shelton, C. D., McNeil, M. B., Early, J. V., Ioerger, T. R. & Parish, T. Deletion of Rv2571c confers resistance to arylamide compounds in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 65, e02334-02320 (2021).
Ligaba, A. et al. Functional, structural and phylogenetic analysis of domains underlying the A l sensitivity of the aluminum-activated malate/anion transporter, T a ALMT 1. Plant J. 76, 766–780 (2013).
Lu, J. et al. Molecular cloning and functional characterization of the aluminum-activated malate transporter gene MdALMT14. Sci. Hortic. 244, 208–217 (2019).
Pohlmann, T., Baumann, S., Haag, C., Albrecht, M. & Feldbrügge, M. A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. Elife 4, e06041 (2015).
Hulbert, S. H., Webb, C. A., Smith, S. M. & Sun, Q. Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312 (2001).
Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20, 116–122 (2004).
Bent, A. F. et al. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860 (1994).
Majeed, S. et al. Role of SNPs in determining QTLs for major traits in cotton. J. Cotton Res. 2, 1–13 (2019).
Li, C. et al. Genome-wide SNP linkage mapping and QTL analysis for fiber quality and yield traits in the upland cotton recombinant inbred lines population. Front. Plant Sci. 7, 1356 (2016).
Liu, R. et al. GWAS analysis and QTL identification of fiber quality traits and yield components in upland cotton using enriched high-density SNP markers. Front. Plant Sci. 9, 1067 (2018).
Byers, R. L., Harker, D. B., Yourstone, S. M., Maughan, P. J. & Udall, J. A. Development and mapping of SNP assays in allotetraploid cotton. Theor. Appl. Genet. 124, 1201–1214 (2012).
Yasir, M. et al. A genome-wide association study revealed key SNPs/genes associated with salinity stress tolerance in upland cotton. Genes 10, 829 (2019).
Sripathi, V. R. et al. Identification of microRNAs and their targets in four Gossypium species using RNA sequencing. Curr. Plant Biol. 14, 30–40 (2018).
Bartel, D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Zhao, T. et al. Identification and profiling of upland cotton microRNAs at fiber initiation stage under exogenous IAA application. BMC Genom. 20, 1–15 (2019).
Akhter, Y. & Khan, J. A. Genome wide identification of cotton (Gossypium hirsutum)-encoded microRNA targets against Cotton leaf curl Burewala virus. Gene 638, 60–65 (2018).
Akmal, M., Baig, M. S. & Khan, J. A. Suppression of cotton leaf curl disease symptoms in Gossypium hirsutum through over expression of host-encoded miRNAs. J. Biotechnol. 263, 21–29 (2017).
Uttara, B., Singh, A. V., Zamboni, P. & Mahajan, R. Oxidative stress and neurodegenerative diseases: A review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 7, 65–74 (2009).
Wieczorek, P. & Obrępalska-Stęplowska, A. Suppress to survive: Implication of plant viruses in PTGS. Plant Mol. Biol. Report. 33, 335–346 (2015).
Barrera-Figueroa, B. E. et al. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 11, 127 (2011).
Yin, Z. et al. Difference in miRNA expression profiles between two cotton cultivars with distinct salt sensitivity. Mol. Biol. Rep. 39, 4961–4970 (2012).
Mondal, T. K. & Ganie, S. A. Identification and characterization of salt responsive miRNA-SSR markers in rice (Oryza sativa). Gene 535, 204–209 (2014).
Shweta, J. A. K. In silico prediction of cotton (Gossypium hirsutum) encoded microRNAs targets in the genome of Cotton leaf curl Allahabad virus. Bioinformation 10, 251 (2014).
Wang, M., Sun, R., Li, C., Wang, Q. & Zhang, B. MicroRNA expression profiles during cotton (Gossypium hirsutum L.) fiber early development. Sci. Rep. 7, 1–13 (2017).
Huffaker, A. & Ryan, C. A. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA 104, 10732–10736 (2007).
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K. & Tran, L.-S.P. Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 64, 445–458 (2013).
Acknowledgements
We are thankful to the International Foundation for Science (IFS), Sweden, for supporting this study under project IFS- I-1-C-6501-1 to AH. This study was funded by NSF awards (IOS-1557796 and IOS-2038872) to MSM.
Author information
Authors and Affiliations
Contributions
A.H. designed research work. A.H., N.A., A.B., J.S., A.R.P., K.I., E.A., A.N., and M.A. performed bioinformatics analysis and experimental work. A.H., A.N., and A.A. wrote the first draft, and M.Z., A.N., M.S.M. reviewed and edited the manuscript. All authors reviewed the final draft and approved it for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hussain, A., Asif, N., Pirzada, A.R. et al. Genome wide study of cysteine rich receptor like proteins in Gossypium sp.. Sci Rep 12, 4885 (2022). https://doi.org/10.1038/s41598-022-08943-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-08943-1
This article is cited by
-
Genome-wide identification and molecular characterization of CRK gene family in cucumber (Cucumis sativus L.) under cold stress and sclerotium rolfsii infection
BMC Genomics (2023)
-
Genome-wide identification of GhRLCK-VII subfamily genes in Gossypium hirsutum and investigation of their functions in resistance to Verticillium wilt
BMC Plant Biology (2023)
-
Comparative transcriptome analysis reveals candidate genes for cold stress response and early flowering in pineapple
Scientific Reports (2023)
-
Genome-wide characterization and expression analysis of cyclic nucleotide-gated ion channels (CNGCs) gene family in Arabidopsis thaliana L. and Helianthus annuus L. for drought stress
Brazilian Journal of Botany (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.