Abstract
Telomerases elongate the ends of chromosomes required for cell immortality through their reverse transcriptase activity. By using the model organism Saccharomyces cerevisiae we defined the order in which the holoenzyme matures. First, a longer precursor of the telomerase RNA, TLC1 is transcribed and exported into the cytoplasm, where it associates with the protecting Sm-ring, the Est and the Pop proteins. This partly matured telomerase is re-imported into the nucleus via Mtr10 and a novel TLC1-import factor, the karyopherin Cse1. Remarkably, while mutations in all known transport factors result in short telomere ends, mutation in CSE1 leads to the amplification of Y′ elements in the terminal chromosome regions and thus elongated telomere ends. Cse1 does not only support TLC1 import, but also the Sm-ring stabilization on the RNA enableling Mtr10 contact and nuclear import. Thus, Sm-ring formation and import factor contact resembles a quality control step in the maturation process of the telomerase. The re-imported immature TLC1 is finally trimmed into the 1158 nucleotides long mature form via the nuclear exosome. TMG-capping of TLC1 finalizes maturation, leading to mature telomerase.
Similar content being viewed by others
Introduction
The protection and maintenance of the ends of linear chromosomes during replication is crucial for dividing eukaryotic cells. Therefore, nucleoprotein structures called telomeres shield the ends of linear chromosomes from double strand break repair activities and thus from end-to-end ligations that would cause massive genome rearrangements and genome instability1,2. These telomeres are the target of a ribonucleoprotein (RNP) complex, called telomerase that protects the chromosome ends from progressive shortening by extending them through its reverse transcriptase activity3. While in most somatic cells telomere shortening occurs after sufficient cell divisions and causes a cell to enter into replicative senescence, stem cells, germ cells, lymphocytes and unicellular eukaryotes like yeast express telomerase to encounter this effect. Remarkably, almost 90% of all cancer cells have re-activated the telomerase to bypass their proliferation limit1,4,5,6. As the production of functional telomerase is crucial for telomere maintenance, it is important to understand its assembly and the order of its step-wise maturation process to realize the underlying control mechanisms that generate functional telomerase.
The telomerase is an RNP complex that contains a long non-coding RNA component, TLC1 (telomerase component 1) composed of 1158 nucleotides in yeast, which functions as template for the synthetic telomere repeats and as a scaffold for the protein components of the holoenzyme, providing the reverse transcriptase activity2,7,8,9 Like mRNAs and most lncRNAs, TLC1 is transcribed in the nucleus by RNA polymerase II (RNAP II) and it is subsequently capped with a monomethyl guanosine (m7G) cap and polyadenylated like mRNAs7,10. However, in contrast to mRNAs, but very similar to snRNAs, Tgs1 generates a 2,2,7-trimethylguanosine (TMG)-cap on TLC1 in the nucleolus, which persists in the mature telomerase7,11,12. Interestingly, TLC1 was described to exist in two forms in the cell, a 1158 nucleotides long mature form and a ~ 1300 nucleotides long precursor. The majority of the TLC1 molecules that are present in cells is the short form and only ~ 10% of the TLC1 RNA is the long polyadenylated form13. This might reflect that approximately 10% of the TLC1 RNAs might be in their maturation phase and 90% are already matured. However, in addition to the cleavage and polyadenylation factor mediated transcription termination (CPF-CF), as used for mRNAs, a study uncovered the ability of the Nrd1-Nab3-Sen1 (NNS) system to terminate TLC1 transcription and it was suggested that this would immediately generate the ~ 1150 nucleotides short form, characteristic of the mature telomerase14,15. If really both termination sites are equally used or if one is preferred over the other under certain conditions is currently unclear. Regardless of the transcription termination pathway used, the primary transcript is always longer than the mature form. Clearly, some of the TLC1 molecules receive a ~ 80 nucleotides long poly(A) tail after transcription, which is removed during the maturation process and the exosome continues the trimming of the pre-TLC1 RNA to 1158 nucleotides7.
In addition to these RNA-specific structural maturation events, TLC1 provides a scaffold for several proteins16. In analogy to the snRNAs, the 3’-end of TLC1 is bound by a heptameric ring, composed of seven Sm-proteins, that encircles and stabilizes TLC112,17,18. Additionally, structurally important proteins as well as regulatory and catalytic factors bind to the TLC1 RNA. One of the stem loops that are formed by TLC1 is bound by the heterodimer Yku70-Yku80, which is important for a persisting nuclear localization through attachment of the telomerase to chromosome ends19,20. The central domain of TLC1, which includes the template domain for reverse transcription, is bound by the catalytic protein subunit Est2 and two accessory factors Est1 and Est32,7. The Est proteins are stabilized by the Pop1, Pop6 and Pop7 proteins, which are also central components in the RNase MRP and the RNase P21,22,23.
Besides TLC1 processing and the protein loading onto this scaffold, this RNA undergoes nucleo-cytoplasmic shuttling7,24. It is exported into the cytoplasm via Mex67-Mtr2 and Xpo1/Crm124. On mRNAs and snRNAs the RNA-contact of Mex67 is mediated by the guard proteins Npl3, Gbp2 and Hrb125,26,27,28. Thus, a similar mechanism is conceivable for TLC1. For Xpo1/Crm1 a physical RNA contact was shown via the cap binding complex (CBC) that interacts with the monomethyl cap25.
After loading of the Est proteins and the Sm-ring in the cytoplasm, TLC1 is re-imported into the nucleus via Mtr1024,29,30. Prevention of shuttling, either by mutations in the export factors or the import factor results in telomere shortening defects and reduced TLC1 levels24,30,31.
For snRNAs the transfer to the cytoplasm was shown to be crucial for the generation of a functional spliceosome, because when not exported, the longer, immature pre-snRNAs can be incorporated into the spliceosome and jeopardize splicing25. Thus, the immediate cytoplasmic drain of these noncoding RNAs is crucial for cell survival and might also be similarly important for the transcribed immature TLC1.
Interestingly, maturation events such as trimming and TMG-capping were suggested to be nuclear events that occur prior to TLC1 shuttling and current models suggest that only the Est proteins and the Sm-ring are loaded in the cytoplasm24,30,31. Therefore, we investigated the maturation steps of TLC1 in detail and uncovered a stepwise process. We found that pre-TLC1 shuttles into the cytoplasm for the loading of the Sm-ring, the Est- and the Pop proteins. The subsequent re-import of the protein bound TLC1 precursor not only requires Mtr10, but also Cse1, a novel import receptor for TLC1. Defects in CSE1 result in amplified Y′ elements and thus elongated telomere ends and destabilized Sm-ring association on pre-TLC1. Thus, proper nuclear re-import of protein-bound pre-TLC1 can only occur in the presence of both, intact Cse1 and Mtr10, which resembles a quality control step in the life cycle of the telomerase RNA. We further show that trimming of TLC1 to the 1158 nucleotides short form is carried out by the nuclear exosome after shuttling. In a final step, TMG-capping of TLC1 completes maturation. In summary we have uncovered that the maturation of TLC1 occurs in a different order than anticipated and includs a cytoplasmic quality control step to ensure proper telomere function.
Results
The Sm-ring is loaded to the precursor of TLC1 in the cytoplasm
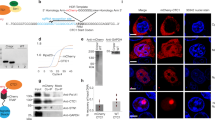
Sm-ring binding to TLC1 is a prerequisite for proper processing of this non-coding RNA, while the contact of the TRAMP-complex and the nuclear exosome prior to the Sm-ring attachment rather initiates full degradation of the transcript17,18. On snRNAs, Sm-ring loading was recently identified to be a cytoplasmic event25. It was shown that newly transcribed snRNAs are immediately exported from the nucleus to prevent an incorporation of the precursor RNAs into the spliceosome, which jeopardizes splicing25. TLC1 receives its Sm-ring also in the cytoplasm (Fig. 1 and Ref.30). To analyze to which form the Sm-ring is loaded, the short 1158 nucleotides long mature form or the longer premature form, we did the following experiments. First, we extracted GFP-tagged Smb1 from cell lysates (Fig. 1A) and analyzed in RNA-co immunoprecipitation (RIP) experiments the general binding of TLC1. Subsequent qPCR experiments showed a clear binding of TLC1 to Smb1, which was normalized to no tag as a negative control and related to 21S rRNA (Fig. 1B). Importantly, when we repeated the experiment in the export mutant mex67-5 xpo1-1 after shifting the cells for 2 h to the non-permissive temperature, which prevents the export of newly synthesized TLC1 molecules, we detected a significantly decreased binding between the Sm-ring component Smb1 and TLC1 (Fig. 1C,D). The decreased binding between Smb1 and U1 served as a control and is in agreement with published data25. These results suggest that the inhibition of the nuclear export of TLC1 prevented the loading of the Sm-ring.
The longer precursor of TLC1 binds the Sm-ring in the cytoplasm. (A) RIP experiments were carried out with GFP-tagged Smb1 from wild type cells. One example IP of Smb1 is shown on western blots. Aco1 served as a negative control. Images were cropped from the original. Original image can be found in Supplementary Fig. S1A. (B) Smb1 binds to TLC1. The Smb1 co-precipitated RNA was used as a template in qPCRs for TLC1. For this (and all following IPs carried out in this study), the amount of the eluted RNA was related to the RNA present in each lysate. Normalization to the mitochondrial 21S rRNA is indicated on the y-axis. n = 4. (C) A western blot shows an example IP of the precipitated Smb1 protein from the RIP experiment shown in (D). Images were cropped from the original. Original image can be found in Supplementary Fig. S1C. (D) Smb1 cannot bind to TLC1 when its nuclear export is prevented. The Smb1 co-precipitated RNA was used as a template in qPCRs for TLC1 in the indicated strains, shifted to 37 °C for 1 h. n = 3. (E) TLC1 accumulates in the cytoplasm of cells in which the Sm-ring is not properly assembled. FISH-experiment of a Cy3-labeled probe (red) targeting TLC1 is shown in wild type cells and a strain in which the Sm-ring is not assembled trough mutation of SMB1 and SMD1 after glucose induced expression repression. The DNA is stained with DAPI. n = 3 (F) Scheme of the ~ 1300 nucleotides long immature TLC1 and the primer positions for the amplification of the total and the immature TLC1 molecules. (G) Western blot showing an example of a Smb1 IP used in (H) and (I) in RIP experiments. Images were cropped from the original. Original image can be found in Supplementary Fig. S1B. (H) Blocking nuclear re-import of TLC1 increases the amount of the total TLC1. RIP experiments and subsequent qPCRs were carried out to show the total TLC1 in the indicated strains. n = 3 (I) Blocking nuclear re-import of TLC1 increases the amount of the immature TLC1. RIP-experiments were carried out and subsequent qPCR results of the immature TLC1 in the indicated strains is shown. n = 3 (J) Western blot showing an example of a Cbp20 IP used in (K) for RIP experiments. Images were cropped from the original. Original image can be found in Supplementary Fig. S1D. (K) Immature TLC1 binds to Cbp20. The Cbp20 co-precipitated RNA was used as a template in qPCRs for immature TLC1. RPL8A served as positive control. n = 3.
This finding is further supported by a fluorescense in situ hybridization (FISH) experiment targeting the TLC1 RNA with a Cy3-labeled probe. TLC1 is mostly detectable in the nucleus of wild type cells (Fig. 1E) and Ref.24. However, in a double mutant of smb1 smd1, which has a defective Sm-ring32, TLC1 mis-localizes to the cytoplasm (Fig. 1E). These findings confirm that Sm-ring loading to TLC1 occurs in the cytoplasm, which was also shown earlier via an inducible TLC1-RNA tagging experiment and is very similar to the cytoplasmic Sm-ring loading onto the snRNAs25,30. Therefore, we investigated the interaction between the Sm-ring and TLC1 in the import receptor mutant mtr10∆. RIP-experiments show an increased binding of total TLC1 (Fig. 1G–I). To get information whether the mature or the immature form of TLC1 accumulates, we also investigated the binding of the precursor of TLC1 to Smb1. A scheme of TLC1 shows that the primers amplifying the unprocessed variant of TLC1 will amplify any generated pre-TLC1, no matter if NNS or CPF-CF terminated (Fig. 1F).
Indeed, as shown in Fig. 1G–I, the binding of the immature TLC1 to Smb1 increased even more in mtr10∆ than that of the total TLC1, suggesting that rather the immature form accumulates. These findings indicate that the Sm-ring is loaded in the cytoplasm to the immature pre-TLC1 and that this form waits for nuclear re-import.
Futhermore, it was shown that the export of TLC1 requires Xpo1 in addition to Mex6724. For snRNA export it was shown that Xpo1 contacts the RNA via the cap binding complex (CBC)25. Since pre-TLC1 contains an m7G cap, it seems likely that this 5’ cap is also bound by CBC. However, this has not been shown so far. Therefore, we carried out RIP experiments with Cbp20 showing that the immature form of TLC1 associates with this CBC-component (Fig. 1J–K). Together, these findings suggest that pre-TLC1 is exported via CBC-bound Xpo1.
Cse1 is a novel nuclear re-import factor for TLC1
Nuclear re-import of TLC1 requires the nuclear import receptor Mtr1029. Interestingly, snRNAs are re-imported via Mtr10 and Cse1, another member of the kayopherin transport factor family25. Interestingly, Cse1 was originally identified as a nuclear export factor for the import adapter protein importin α, encoded by SRP133,34. However, recently we have shown that in addition to its exporting function, Cse1 supports the import of Sm-ring-containing snRNAs, which thus resembles an important control step for its maturation, because the importer only interacts with Sm-ring containing snRNAs25. As the Sm-ring is also loaded onto TLC1 in the cytoplasm, it seems reasonable to assume that Cse1 would also participate in its nuclear re-import. Therefore, we localized TLC1 in the cse1-1 mutant that was shifted to 16 °C, as it is cold sensitive. We found that while TLC1 localized mainly to the nucleus in wild type cells, it was distributed throughout the cytoplasm in the cse1-1 mutant (Fig. 2A, Supplemental Fig. S1), similar to mtr10∆ and yku70∆ strains (Fig. 2A) and Refs.24,29. Since Mtr10 and Cse1 both contribute to the nuclear import of TLC1 and snRNAs, we created a double mutant and assayed its growth. Although the single mutants already showed growth defects in comparison to wild type, the double mutant grew even less, indicating a genetic interaction between these import receptor genes (Fig. 2B). Subsequent FISH experiments with the double mutant also showed a cytoplasmic accumulation, similar to that observed in the single mutants (Fig. 2A).
Cse1 is a novel nuclear import factor for TLC1. (A) TLC1 mis-localizes to the cytoplasm in cse1 mutants. A FISH-experiment for TLC1 (red) is shown in the indicated strains that were shifted to the indicated non-permissive temperatures. n = 3. Overview of the cells is shown in Supplementary Fig. S2A and negative controls in Supplementary Fig. S2B,C. (B) CSE1 and MTR10 genetically interact. Serial dilutions of the indicated strains were spotted onto full medium agar plates and incubated at the indicated temperatures. (C) RIP experiments were carried out with GFP-tagged Mtr10 and Cse1 from wild type cells. One example IP is shown on a western blot. Zwf1 served as a negative control. Images were cropped from the original. Original image can be found in Supplementary Fig. S2D. (D) Cse1 binds to TLC1. The Cse1 co-precipitated RNA was used as a template in qPCRs for total TLC1. n = 4 Mtr10 served as a positive control. n = 4 (E) RIP experiments were carried out in the indicated mutant strains. A western blot of an example IP is shown. Aco1 served as a negative control. Images were cropped from the original. Original image can be found in Supplementary Fig. S2E. (F) Binding of total TLC1 to Smb1 decreases in cse1-1 and cse1-1 mtr10∆ cells. RIP experiments and subsequent qPCRs were carried out. n = 4. (G) Binding of immature TLC1 to Smb1 increases in cse1-1 and cse1-1 mtr10∆ cells. RIP experiments and subsequent qPCRs were carried out. n = 4. (H) The ratio of the Smb1-bound immature versus total TLC1 increases in cse1-1 mtr10∆. The ratios were calculated from the experiments shown in F and G. n = 4. (I–K) The immature form of TLC1 accumulates in nuclear re-import mutants. The RNA was isolated from the indicated strains after a 1 h 15 min temperature shift to the non-permissive temperature. Subsequent qPCRs were carried out with primers that detect either all TLC1 forms (total TLC1) (I) or only the unprocessed form (immature TLC1) (J). (K) The ratio of immature to total TLC1 increases in the import mutants. The ratios were calculated from the experiments shown in (I) and (J). mtr10-1 n = 3. cse1-1 n = 6, mtr10∆ n = 7 and cse1-1 mtr10∆ n = 5.
To verify binding of TLC1 to Cse1, we carried out RIP experiments, in which we precipitated GFP-tagged versions of the import receptors Cse1 and Mtr10, the latter of which served as a positive control (Fig. 2C,D). We found that the binding of TLC1, most likely its immature form to both import receptors was significantly enriched, similar to the binding of the snRNAs U1 and U5 (Fig. 2D) and Ref.25. A 1.5 to twofold increase of the binding, although significant, might on first sight not seem to be very strong, but considering that the binding of both import receptors depends on the small GTPase Ran the low value is understandable. For import-cargo complex formation Ran must be in its GDP bound state. This is favored in the cytoplasm by the presence of the GTPase activating protein RanGAP1 and its co-factor RanBP135,36,37. Cargo release from the import receptors occurs in the nucleus where RanGTP is present and the nucleotide exchange on Ran occurs with the nuclear exchange factor Prp20 (human RCC1)37. Upon cell lysis for the RIP experiment, both compartments get mixed up and pre-formed nuclear import complexes are attacked and disassembled through RanGTP. An additional difficulty is the low copy number of TLC1 with ~ 10 to ~ 30 copies per haploid cell7, 38,39. However, despite these difficulties, we were able to detect binding of TLC1 to both import factors, Mtr10 and Cse1 (Fig. 2C,D).
Analogous to mtr10∆ (Fig. 1H,I) we investigated the interaction of TLC1 and the Sm-ring in cse1-1 and the cse1-1 mtr10∆ double mutant. Interestingly, in contrast to mtr10∆ where we observed an increased protein-RNA interaction, defective cse1 leads to a decreased TLC1-Sm-ring binding for both, immature and mature TLC1, suggesting a stabilizing function of Cse1 for Sm-ring formation. Reduced Sm-ring formation in cse1-1 did not affect the relative level of TLC1 (Fig. 2I–K), suggesting that pre-TLC1 might still be protected by other proteins. In the double mutant, in which both import receptors are missing, the Sm-ring binding is even further reduced to ~ 15% (Fig. 2E–G). In contrast to cse1-1, mutation in MTR10 lead to reduced cellular levels of TLC1 to about 50%, which is also the case in the double mutant cse1-1 mtr10∆ (Fig. 2I–K). Our data suggest that although TLC1 is mis-localized to the cytoplasm in both import factor mutants and both factors contribute to the re-import of this RNA, their functions differ in some aspects. Mtr10 seems to be the main import factor, as Sm-ring bound RNA accumulates in mtr10∆ and thus reduces the overall level of TLC1 (Fig. 1H,I). In contrast to this, Cse1 seems to stabilize the Sm-ring formation on pre-TLC1 and mutants therefore rather delay maturation and in turn nuclear re-import. Remarkably, when we compare the binding of the Sm-ring to the immature TLC1 in the different mutants, we still detect increased amounts (Fig. 2H), suggesting that this form is trapped in the cytoplasm and that Cse1 participates in nuclear import.
Together, these findings identify Cse1 as a novel import receptor that is also required for Sm-ring loading for TLC1 and show that optimal nuclear re-import of this a large RNP relies on more than one import receptor.
Mutation of CSE1 leads to amplified Y′ elements
Prevention of nuclear TLC1 shuttling leads to shortened telomeres as seen in the export receptor Mex67 and Xpo1 as well as in the import receptor Mtr1024,31 (Fig. 3C). To investigate whether such defects would also be visible in the new import factor Cse1, we carried out southern blot analyses. Strikingly, we detected no shorter telomere ends in cse1-1, but instead highly amplified subtelomeric Y’ elements (Fig. 3A,B). Usually, cells that lack the telomerase or components of it undergo senescence when telomeres become critically short. The senescence phenotype is only temporary and most of these cells lose their viability, but a small percentage of cells overcome this phenotype by lengthening telomeres through homologous recombination, termed Type I and Type II survivors40,41. Type I survivors depend on Rad51 and exhibit highly amplified subtelomeric Y’ elements42, which are clearly visible in cse1-1 cells (Fig. 3B,C). In contrast to the Type I survivors, cse1-1 mutants have no shortened telomeres. It’s phenotype rather resembles a mutant in which telomere cap binding proteins, such as Rap1, are depleted.
Mutation of CSE1 results in Y′ element amplification. (A) Scheme of the Y′-amplification and location of the probe that was used for the southern blot. (B) The cse1-1 mutation leads to the amplification of the telomeric Y′ element. XhoI digested genomic DNA of the indicated strains was used for the southern blot. The chromosome ends were detected with a digoxygenin labeled probe (A). Original image can be found in Supplementary Fig. S3A. (C) Mutation of CSE1 leads to the amplification of of the telomeric Y′-element. Mutation of MTR10 leads to shortend telomere ends. XhoI digested genomic DNA of the indicated strains was used for the southern blot. The chromosome ends were detected with a digoxygenin labeled probe (A). Image was cropped from the original. Original image can be found in Supplementary Fig. S3B. (D) Plasmid encoded CSE1 recues the cse1-1 phenotype. XhoI digested genomic DNA of the indicated strains was used for the southern blot. The chromosome ends were detected with a digoxygenin labeled probe (A). Original image can be found in Supplementary Fig. S3C. (E) Mutation of CSE1 did not lead to a senescence phenotype. n = 1. (F) TLC1 localization is not altered in the srp1-31 mutant. FISH experiments show no mis-localization of TLC1 in srp1-31 after shift to 37 °C for 1 h. n = 3. (G) Cdc13 localization is not altered in the cse1-1 mutant. Immunofluorescence experiments show no mis-localization of Cdc13 in cse1-1 after shifting the cells to 16 °C for 1 h and 15 min. n = 3. (H) Rap1 localization is altered in the cse1-1 mutant. Immunofluorescence experiments shows nucleolar mis-localization of Rap1 in cse1-1 at the permissive temperature and a cytoplasmic mis-localization after shifting the strain to 16 °C for 1 h and 15 min. n = 3. (I) Model for Y′ element amplification in the cse1-1 mutant. In wild type cells (upper panel) A, long and capped telomeres are usually not elongated via the telomerase. To short telomeres, B, telomerase is recruited and telomeres are elongated. In the cse1-1 mutant (lower panel) defects in Rap1 import lead to unprotected telomeres, which triggers the Y′ amplification via homologous recombination.
In addition, we analyzed whether the telomeric phenotype in the cse1-1 mutant can be reversed by introduction of a plasmid containing wildtype CSE1. Already after one restreak, which equals approximately 25 generations the Y' amplification band disapeared, indicating that the phenotype is based on the cse1 mutation (Fig. 3D). Furthermore, it can be seen that upon re-streaking of the tlc1∆ strain on a plate, Type I survivors are generated43 that have a short terminal fragment and Y′ amplification (Fig. 3D). This switch to homologues recombination in the tlc1∆ goes along with a kink in the growth curve (Fig. 3E), which is absent in cse1-1 cells.
Cse1 is also involved in the recycling of importin alpha, encoded by SRP1 and cse1-1 leads to an accumulation of Srp1 in the nucleus33. It was reported earlier that Srp1 might be involved in the nuclear localization of Est144. Therefore, we examined whether TLC1 mis-localizes in an srp1-31 mutant. FISH experiments show no mis-localization of TLC1 in srp1-31 (Fig. 3F, Supplementary Fig. S2A,B).
Since short and uncapped telomeres are prone to recombination and often result in Y′ amplification we wondered whether an altered telomere cap structure in the cse1-1 mutant might elicit the observed phenotype. Thus, we analyzed the localization of two important telomere cap components. While the localization of Cdc13 was not affected by the cse1-1 mutation, Rap1 was mis-localized to the nucleolus at the permissive growth temperature of cse1-1 and to the cytoplasm at the non-permissive temperature (Fig. 3G,H, Supplementary Fig. S2C,D). The resulting Rap1 depletion in the nucleus at telomeres might be the cause of the Y′ amplification in cse1-1.
Pop protein loading onto TLC1 occurs in the cytoplasm
After knowing the nuclear re-import requirements for TLC1, we asked, where the proteins of the RNP are loaded onto TLC1 in the cell. We have suspected earlier that Est protein binding occurs in the cytoplasm, as the Est proteins accumulate in this compartment in a tlc1∆ mutant24. Reassuringly, we also detect a cytoplasmic mis-localization for Est1-GFP in the new TLC1 re-import factor mutant cse1-1 (Fig. 4A,B).
Loading of the Est and Pop proteins occurs in the cytoplasm. (A) Nuclear GFP-tagged Est1 mis-localizes to the cytoplasm in cse1-1 cells. n = 3. (B) A wild type strain is shown without a GFP tagged protein with identical microscopy settings as shown in (A). (C) Nuclear Pop1 localization is disturbed in mutants affecting TLC1 localization. n = 3. (D) Western blot of the Pop1-GFP IP is shown in the indicated strains. Grx4 served as a negative control. n = 3. Image was cropped from the original. Original image can be found in Supplementary Fig. S4A. (E) Pop1 binding to TLC1 is decreased in an export mutant. The Pop1-bound RNA from the several IP, one of which is shown in D, was analyzed in qPCRs n = 3. (F) Western blot of the Pop1-GFP IP is shown in the indicated strains. Grx4 served as a negative control. n = 3. Image was cropped from the original. Original image can be found in Supplementary Fig. S4A. (G) Pop1 binding to TLC1 is increased in an import mutant. The Pop1-bound RNA from the several IP, one of which is shown in F, was analyzed in qPCRs. n = 3.
For the Pop proteins the place of loading onto TLC1 is currently unknown. As the proteins are mostly localized to the nucleolus and the nucleoplasm45, it is possible that they are loaded after TLC1 has completed its cytoplasmic phase and returned to the nucleus. The function of the Pop proteins in TLC1 maturation is to stabilize and recruit Est1 and Est2 within the holoenzyme22,23. Therefore, it is also conceivable that they are loaded to TLC1 in the cytoplasm, before the Est proteins were properly attached. Importantly, Pop1 can only be stably associated with TLC1 when the heterodimer Pop6 and Pop7 was attached to the RNA46. Thus, Pop1 binding is the final step in the loading of the Pop protein complex. We localized Pop1 in the TLC1 nuclear import- and export mutants and in tlc1∆ and found a clear cytoplasmic mis-localization of the protein, even though we detect its localization in wild type not as strict nuclear as published earlier with the identical constructs45 (Fig. 4C). Interestingly, also mutations in the Pop proteins were shown to increase the cytoplasmic presence of TLC121, which supports a model in which loading of the Pop proteins might occur in the cytoplasm. Thus, in situations, in which TLC1 is not present in the cytoplasm, such as in tlc1∆ or in the export mutant mex67-5 xpo1-1, the Pop proteins accumulate in the cytoplasm and most likely do not bind TLC1. However, in the import mutants, one would expect that the proteins were loaded onto TLC1, but cannot be imported. To get additional support for a cytoplasmic loading and to find out whether Pop1 is loaded onto the long immature TLC1 we carried out RIP experiments, with strains that contained endogenously tagged POP1-GFP expressed from its own promoter to avoid overexpression. Clearly, we found a decreased binding of this RNA in the export mutant (Fig. 4D,E), while an import block resulted in an increased binding of the immature TLC1 to Pop1 (Fig. 4F,G), suggesting that the loading of the Pop proteins is indeed a cytoplasmic event.
Mtr10 contacts TLC1 at the Sm-ring for nuclear re-import
Cse1 can mediate the nuclear import of snRNAs and TLC1 only after the Sm-ring has properly assembled25 (Fig. 1E). An interaction between the Sm-ring and Cse1 has been shown before25. Their interaction thus resembles a quality control step to allow only nuclear entrance of the Sm-ring-bound TLC1 RNP. It seems possible that also Mtr10 may contact TLC1 only after proper protein loading has occurred. To analyze potential complex formations, we carried out co-immunoprecipitations (co-IPs) of Mtr10 with Pop1 and Est1. However, none of these proteins physically interacted with Mtr10 (Fig. 5A,B).
Both nuclear import receptors contact TLC1 at the Sm-ring. (A) Mtr10 does not interact with the Pop protein complex. A western blot of co-IPs of GFP-tagged Pop1 with myc-tagged Mtr10 is shown. Nop1 served as a washing control n = 3. Image was cropped from the original. Original image can be found in Supplementary Fig. S5.1A. (B) Mtr10 does not interact with the Est protein complex. A western blot of a co-IP of GFP-tagged Mtr10 with myc-tagged Est1 is shown. Nop1 served as negative control. n = 3. Image was cropped from the original. Original image can be found in Supplementary Fig. S5.2A. (C) Mtr10 does not interact with the Yku protein complex. A western blot of a co-IP of myc-tagged Mtr10 with Yku70 is shown. n = 3. Image was cropped from the original. Original image can be found in Supplementary Fig. S5.3A. (D) Mtr10 interacts with the Sm-ring. A western blot of a co-IP of GFP-tagged Smb1 with myc-tagged Mtr10 is shown. Aco1 served as a negative control. n = 3. Image was cropped from the original. Original image can be found in Supplementary Fig. S5.3B.
It is currently unclear whether the Yku-heterodimer is loaded in the cytoplasm. The Yku70-Yku80 heterodimer binds to a 48 nucleotides long stem loop of TLC1 and is required for the nuclear localization of this RNA19,29,47 (Fig. 2A). Therefore, we investigated a potential binding between Mtr10 and Yku70 in co-IP experiments, but we did not detect an interaction (Fig. 5C).
Finally, we investigated, whether Mtr10 might, like Cse1, also interacts with the Sm-ring in co-IPs and indeed, a specific band of Mtr10 was detectable in the eluates of the Smb1-IPs (Fig. 5D). These findings indicate that both import receptors interact with the Sm-ring to support nuclear import of the matured TLC1 RNP.
Processing of TLC1 occurs in the nucleus after re-import of the protein bound RNP
The longer pre-TLC1 is trimmed by the exosome up to the Sm-ring, resulting in a 1158 nucleotides long form7. Our studies in the re-import mutants suggest that trimming occurs after shuttling, because of an increased Smb1-binding to the immature precursor RNA of TLC1 (Figs. 1, 2H). Moreover, mutants that accumulate TLC1 in the cytoplasm accumulate immature TLC1 at decreasing total TLC1 levels (Fig. 2I–K), suggesting that the TLC1 RNA is not trimmed before its journey through the cytoplasm.
To further analyze this point, cytoplasmic fractionation experiments were carried out. Compared to wild type cells, more immature than total TLC1 accumulates in the cytoplasm of the import factor mutants (Fig. 6A,B). These findings suggest that the immature form shuttles into the cytoplasm and trimming occurs afterwards by the nuclear exosome. Secondly, we assayed the TLC1-forms in the mex67-5 nuclear export mutant. We detected a decrease of total TLC1, which is in agreement with earlier findings in which Mex67 was suggested to protect TLC1 from its degradation30. However, in our experiments we still see a two-fold increase of the immature TLC1 in mex67-5 mutants, which suggests that immature TLC1 is not immediately degraded when Mex67 is missing (Fig. 6E). Possibly, other proteins such as the guard proteins, one of which is Npl3, that recruit Mex67 protect the immature TLC1 from being degraded while waiting for export. To test whether Npl3 binds TLC1, we precipitated the protein and found TLC1, in particular the immature form to bind Npl3 (Fig. 6C,D). To address whether the trimming to the mature TLC1 occurs via the nuclear exosome, we analyzed TLC1 in rrp6∆. Its deletion leads to an approximately two-fold increase of the total TLC1, which includes the mature form and the immature form, and a ~ sevenfold increase of the immature form of TLC1 (Fig. 6E), indicating that the immature form is trimmed in the nucleus. Remarkably, although the RNA is less stable in mex67-5 mutants, the immature TLC1 still increased five-fold in the double mutant rrp6∆ mex67-5, suggesting (a) that its trimming occurs in the nucleus and (b) that the immature form is still protected without Mex67. Furthermore it has to be noted, that in contrast to Vasianovich and colleagues, we find no rescue of the decreased TLC1 in the double mutant rrp6∆ mex67-5 as detected by Northern blot30. Since in our experiments we applied the highly sensitive qPCR method, we were able to detect an increase of the immature TLC1 when its export is blocked in mex67-5, despite the overall decrease of the total TLC1 in both mutants (Fig. 6E).
Processing and TMG-capping of TLC1 occurs in the nucleus upon TLC1 re-import. (A) Immature TLC1 accumulates in import factor mutants. A Western blot of a nucleo-cytoplasmic fractionation experiment is shown for the indicated strains. The nuclear Nop1 protein and the cytoplasmic Zwf1 protein served as controls for successful isolation of the cytoplasmic fraction. Strains were shifted to 16 °C for 1 h 15 min prior to the experiments. n = 5. Image was cropped from the original. Original image can be found in Supplementary Fig. S6A. (B) qPCRs of the fractionation experiments shown in (A) reveals an accumulation of the immature TLC1 in the import factor mutants at the permissive termperature. n = 5. (C) Western blot showing an example of a Npl3 IP used in (D) in the RIP experiments. Image was cropped from the original. Original image can be found in Supplementary Fig. S6B. (D) TLC1 binds to Npl3. The Npl3 co-precipitated RNA was used as a template in qPCRs for TLC1 and Hem15 as positive control. n = 4. (E) Trimming of TLC1 is mediated by the nuclear exosome after nuclear re-import. The RNA was isolated from the indicated strains after shifting them to 37 °C for 2 h. Subsequently, qPCRs were carried out that determined the amount of the total TLC1 and its immature, longer precursor. Mex67-5 and rrp6∆ mex67-5 n = 4. Rrp6∆ n = 8. (F) TMG-capping occurs after nuclear TLC1 re-import. The amount of TMG-capped TLC1 was determined by TMG-co-IPs and subsequent qPCRs from the indicated strains. tgs1∆ served as a positive control and the black line indicates the amount of the precipitated non-TMG-capped RNAs resembling the base line for TMG-capping. Tgs1∆ and mtr10∆ n = 4. Cse1-1 and cse1-1 mtr10∆ n = 3.
Taken together, these finding show that trimming of the immature TLC1 requires nuclear export and that correct trimming occurs only after Sm-ring loading, which seems logic, because the Sm-ring was shown to protect TLC1 from full degradation17. Our data furthermore suggest that the immature TLC1 is protected from degradation by Mex67 binding and its immediate export activity into the cytoplasm. But they also suggest that additional proteins, like Npl3, protect the immature TLC1 from degradation, possibly the guard proteins that recruit Mex67 after quality control, as the immature form increases in the nucleus of mex67-5 mutants (Fig. 6D,E).
Another modification of TLC1 is the TMG-capping at its 5’-end in the nucleolus by Tgs111,48. To investigate, whether this trimethylation occurs before or after shuttling we carried out RIPs experiments with a TMG-cap specific antibody. Subsequently, we carried out qPCRs to detect TLC1. Compared to wild type, we found an approximately 30% decrease in the level of the TMG-capped TLC1 in a TGS1 deletion strain, which resembles the zero-line for the unspecific binding of this antibody, because Tgs1 is the only trimethyltransferase in yeast. Clearly, also in the single import mutants mtr10∆ and cse1-1 as well as in the double mutant we found a reduction, which was around 30% (Fig. 6F). These findings indicate that TMG-capping occurs after shuttling.
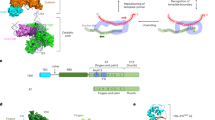
Taken together, we have uncovered a stepwise maturation cycle for the telomerase, which is summarized in Fig. 7. Maturation begins with the nuclear export of the longer pre-TLC1 into the cytoplasm by Mex67, probably assisted by Npl3 and Xpo1 that binds to the CBC. In the cytoplasm not only the Sm-ring is loaded, but also the Est- and the Pop proteins attach to the immature TLC1. Its successful assembly allows Mtr10 and Cse1 to bind and re-import pre-TLC1 into the nucleus. Subsequently, trimming of the immature form into the mature 1158 nucleotides long form is mediated by the nuclear exosome and TMG capping finally occurs in the nucleolus by Tgs1.
Model for the compartmental stepwise maturation of TLC1. The longer precursor of TLC1 is transcribed in the nucleus and immediately exported into the cytoplasm upon binding of the export receptor Mex67-Mtr2 and the karyopherin Xpo1/Crm1, the latter of which interacts with CBC bound m7G-caps. Upon export, Mex67 and Xpo1/Crm1 are displaced and the Sm-ring, the Est and the Pop proteins assemble on the immature TLC1 in the cytoplasm. Subsequently, this RNP is re-imported back into the nucleus via Mtr10 and Cse1, both of which contact TLC1 via its Sm-ring. Thus, nuclear re-import can only occur after RNP formation including the Sm-ring and therefore resembles an important quality control checkpoint. In the nucleus, the import receptors dissociate and TLC1 is trimmed up to the Sm-ring by the nuclear exosome. Finally TMG-capping occurs in the nucleolus assisted by Smb1, which interacts with Tgs1. This step terminates shuttling, because export receptors cannot be loaded anymore. The matured holoenzyme subsequently acts in telomere maintenance.
In our compiled investigation we have ordered the maturation process of TLC1 and show the order of events from nuclear export of the premature transcript to the binding of the Est- and Pop proteins and the Sm-ring in the cytoplasm to its trimming and TMG-capping after re-import into the nucleus. We show that the Sm-ring binding resembles a quality control step that enables subsequent movement of the RNP into the nucleus. Thus, the step-wise maturation of TLC1 is controlled by its compartmental shuttling.
Discussion
Functional telomerases are important to prevent the repetitive shortening of the DNA ends after each round of replication for recurrent cell divisions. Due to the fact that these holoenzymes undergo a stepwise assembly, a danger is that incompletely assembled telomerases interfere with the process of telomere elongation. In the worst case, these immature telomerases could, due to their restricted or absent functionality, prevent telomere maintenance. To avoid such a scenario, a mechanism has developed, in which the RNA scaffold, TLC1 in yeast, is exported into the cytoplasm via the export receptors Mex67-Mtr2 and Xpo1/Crm124,29. While Xpo1/Crm1 contacts the CBC, which is bound to the monomethyl cap of the RNA-polymerase II transcribed RNA, Mex67-Mtr2 was speculated to contact the RNA via the adapter proteins Npl3, Gbp2 and Hrb1, because such contact is observed for mRNA and snRNA export25,26,27,28.
Binding of the Est proteins to TLC1 and association of the Sm-ring were suggested to be cytoplasmic events, while TMG-capping and trimming of TLC1 were suggested to occur in the nucleus prior to shuttling24,29,30. Up to date, the place of the Pop protein loading onto TLC1 was unknown, but suspected to occur in the cytoplasm, because mutants of POP1 and POP6 accumulated TLC1 in the cytoplasm21. We collected additional evidence for a cytoplasmic Pop protein loading and analyzed the localization of Pop1 in the import factor mutants. We found a cytoplasmic accumulation of Pop1 in the mutants (Fig. 4C), supporting the idea of their cytoplasmic loading and supporting the newly discovered role of Cse1 in TLC1 nuclear import. Finally, we could confirm this result through RIP experiments in which we detected a decreased TLC1 binding to Pop1 in mex67-5 and an increased binding in the import mutant cse1-1 (Fig. 4E,G). As the function of the Pop proteins is to stabilize the Est proteins on the RNA and to support the RNP structure22,23 their cytoplasmic loading seems to be a final step before re-import.
Another important event is the loading of the Sm-ring that limits the trimming of the TLC1 precursor, which was recently shown to occur in the cytoplasm, similar to the Sm-ring loading onto snRNAs25,30. However, in the interesting study of Vasianovich and colleagues a modified TLC1 that contained several MS2 stem loops was used, which could have an influence on its localization. In addition, they could not distinguish which form of TLC1 is accumulating in the cytoplasm. In our study we used probes and primers that (1) detect the unmodified natural TLC1 and (2) selectively detect the immature pre-TLC1. With this method we confirmed the data of Vasianoivich and colleagues and show that the Sm-ring loading is a cytoplasmic event. But in addition, we have shown that the Sm-ring and the Pop proteins associate with the immature precursor of TLC1 in the cytoplasm (Figs. 1I, 2H, 4G) and that trimming occurs after Sm-ring loading and after re-import into the nucleus (Figs. 1I,K, 2H,K, 4E,G, 6E).
It was shown that the karyopherin Cse1 contacts the Sm-ring and particularly Smb1 for nuclear re-import of the snRNAs25. Interestingly, we also detected a binding between the Sm-ring and the other import factor, Mtr10 (Fig. 5D). This finding is important for three reasons. First, besides Mtr10, Cse1 is another import factor that facilitates the transit of this huge RNP through the NPC. The more transport factors are involved in transporting large particles, the better the transit is working. This is supported by the additive effect for the mis-localization of TLC1 seen in the combined import factor mutants (Fig. 6A,B). Secondly, a potential failure of one of the import receptors does not fully prevent TLC1 re-import. Third, nuclear re-import supported by Mtr10 and Cse1 can only occur on TLC1 molecules that have received the Sm-ring, which resembles an elegant way of cellular quality control for this important maturation step of the telomerase. For this, Cse1 might be more important because we detect a decreased binding of TLC1 to Smb1 in cse1-1 cells, even though we see no drastic change in the overall amount of TLC1 (Fig. 2F–K). Together, these findings argue for a control step in the RNP formation, which prevents that faulty RNPs can enter the nucleus and jeopardize telomere function. However, we cannot exclude that another protein is involved in its stabilization, which is dependent on Cse1 transport. In every sense, Sm-ring loading is an essential step in maturation and allows nuclear re-import of pre-TLC1.
Interfering with the shuttling of TLC1 either by mutation of the export factors Mex67 and Xpo1 or the import receptor Mtr10 was shown to result in telomere shortening defects24,31. Interestingly, we did not detect any telomere shortening defects and no senescence phenotype for the novel TLC1 re-import factor mutant cse1-1 (Fig. 3B,E). Instead, mutation of CSE1 resulted in an Y′ element amplification (Fig. 3A,B). In addition to a functional telomerase an altered telomere cap structure can also impact the telomere length homeostasis. Cap-protecting proteins are the dsDNA binding protein Rap1 and the ssDNA binding protein Cdc13. Since Rap1 is either interacting with the Sir complex on centromere proximal regions or with the Rif1 and Rif2 proteins on centromere distal regions, one might speculate that either the nuclear import of Rap1 itself, or one of its tethering proteins might depend on Cse1 for nuclear import. Indeed, we found a cytoplasmic mis-localization of Rap1 in cse1-1 that supports this idea. Interestingly, Rap1 was also mis-localized at the permissive temperature of cse1-1. However, in contrast to the non-permissive temperature, it was found in the nucleolus, not in the cytoplasm, which might also suggest that its telomere tethering factors were absent, possibly not properly imported by Cse1.
Even though Cse1 mediates the re-import of TLC1, mutation of this import factor additionally impacts proper telomere cap formation, which might elicit the observed phenotype. Here, the importer seems to have a function in transporting Rap1 or a factor that prevents proper Rap1 localization on chromosome ends.
Furthermore, one can speculate that either Rad51 itself, or factors of the Rad51 homologous recombination pathway might not be imported in mtr10∆, so that mtr10∆ mutants cannot use homologous recombination for telomere maintenance but rather show a shortening of the telomeres.
Moreover, we have excluded an involvement of Srp1 in the localization of TLC1. Srp1 was suggested to mediate the nuclear localization of Est1 and since Srp1 mis-localizes in the cse1-1 mutant an influence also on TLC1 was conceivable33,44. However, TLC1 and thus telomerase localization does not appear to be dependent on Srp1, as mutation of SRP1 does not lead to a mis-localization of TLC1 (Fig. 3F).
At steady state about 90% of TLC1 are present in the mature shorter form which is present in the functional telomerase and only a small fraction is present as the polyadenylated longer form9,13. Older model discuss the CPF-CF terminated poly(A)+ form as a precursor of the mature form13, but more recent studies suggest that the NNS terminated form might be the prominent one15. Therefore, it is currently unclear how much of the TLC1 originates from NNS or CPF-CF termination. However, since both versions (a) need to adopt the Sm-ring for general protection, which was shown to be a cytoplasmic process25 and (b) total TLC1 significantly decreases if shuttling is prevented (Fig. 2I) and (c) leads to defects of the telomeric structure (Fig. 3) and Ref.24, shuttling of any immature form seems highly relevant.
It was unclear when trimming occurs. Earlier studies indicated that mutations in the nuclear component of the exosome resulted in a strong increase of TLC117. However, the experimental set up did not allow to distinguish whether the exosomal trimming occurs before or after shuttling. To determine the time and place of the TLC1 trimming, we first showed that the immature form accumulates in nuclear re-import mutants and in mutants that are defective in the Sm-ring assembly (Figs. 1 and 2I–K). Additionally, we have shown by nucleo-cytoplasmic fractionation experiments that the immature form accumulates in cytoplasm in the import factor mutants (Fig. 6B), which indicates that trimming occurs after re-import. Furthermore, mutation of RRP6 resulted in a strong increase of the immature pre-TLC1 (Fig. 6E), suggesting that the nuclear exosome is responsible for trimming after shuttling. This finding also suggests that the immature form is still protected without Mex67. One possibility is that the Mex67-interacting and RNA-binding proteins Npl3, Gbp2 and Hrp1 might guard TLC1. And indeed, we have shown that TLC1 and especially the immature TLC1 is bound by Npl3 (Fig. 6D). Co-transcriptionally loading of Npl3 prior to binding of Mex6749,50 provides coverage and thus protection of the RNP.
The finding, that Sm-ring binding occurs before trimming seems logical, because unprotected RNA is an ideal substrate for the exosome18. Interestingly, preventing nuclear export by mutation of MEX67 leads to a decrease of TLC1 (Fig. 6E and Ref.30). During the short period of time in which the immature TLC1 RNA is usually present in the nucleus before Mex67 and Xpo1 export it into the cytoplasm, the nuclear exosome is not able to attack the RNA sustainably. Thus, Mex67 cannot be the only protecting RNA-binding protein, because even in mutants of this export receptor we detect a ~ twofold and a ~ fivefold increase of the immature TLC1 in mex67-5 and rrp6∆ mex67-5, respectively, as compared to wild type. Possibly this is achieved by guard proteins such as Npl3 that recruit Mex6728,50.
Our finding that the Sm-ring loading occurs in the cytoplasm prior to TMG-capping is in agreement with other studies that have shown that the Sm-ring is crucial for trimethylation of the cap51. Furthermore, we have shown earlier that snRNAs are retained in the nucleolus when Smb1 is depleted that showed that an intact Sm-ring is required for efficient trimethylation and subsequent nucleolar release into the nucleoplasm25. It is well conceivable that the formed Sm-ring assists TMG capping, as it interacts with Tgs1 both, in vivo and in vitro11,25.
Importantly, snRNA nucleo-cytoplasmic shuttling was suggested to be terminated by trimethylation of the 5’ cap25. The export receptor Xpo1 interacts with the 5’ cap binding complex CBC, in particular with Cbp8025. This might be similar for TLC1, because pre-TLC1 contains an m7G cap, which is bound to CBC (Fig. 1K) and TLC1 interacts with Xpo124. After receiving the TMG cap, CBC might not be bound and Xpo1 then unable to interact. Since the other export receptor, Mex67, is already displaced at the NPC when entering the cytoplasm52,53, Xpo1 is the only transport factor left for a repeated snRNA export and TMG capping could close this option. Therefore, it seems logic that TMG capping on TLC1 also occurs after shuttling and maturation and we could indeed show that preventing nuclear re-import of TLC1 results in the accumulation of m7G, but not TMG capped RNAs (Fig. 6F).
Together, our analysis uncovered the steps in which telomerase maturation takes place (Fig. 7) and identified that proper cytoplasmic assembly of the telomerase is prerequisite for nuclear re-import and thus represents a quality control check point in the life cycle of this holoenzyme. Any way of interfering with the compartmental maturation process inevitably leads to defects in telomere biology (Fig. 3) and19,21,24,29,31,54,55, reflecting the importance of this step-wise process.
Experimental procedures
Yeast strains, plasmids and oligonucleotides
All yeast strains used in this study are listed in the Supplementary Table S1, oligonucleotides in Supplementary Table S2 and plasmids in Supplementary Table S3. Plasmids and yeast strains were generated by conventional methods.
Fluorescent in situ hybridization experiments (FISH)
The experiments were essentially carried out as described25. RNA probes were with Cy3-labeled oligonucleotides (Sigma), which are listed in Supplementary Table S3. Cells were grown to mid log phase (1 × 107 cells/ml) prior to temperature shift to 37 °C 1 h or 2 h or to 16 °C for 1 h 15 min (Figs. 2A, 3F, Supplemantry Fig. S2A–C, Supplemantry Fig. S3D,E). For Sm-ring dependent localization studies, cells were grown to log phase in YP medium containing 2% galactose. Afterwards 4% glucose was added and cells were incubated at 25 °C for 2 h (Fig. 1E). Samples were fixed by adding formaldehyde to a final concentration of 4% for 45 min at room temperature. Cells were spheroplasted by adding zymolyase, subsequently permeabilized in 0.1 M potassium phosphate buffer pH 6.5, 1.2 M sorbitol, 0.5% Triton® X-100, pre-hybridized with Hybmix (50% deionized formamide, 5 × SSC, 1 × Denhardts, 500 μg/ml tRNA, 500 μg/ml salmon sperm DNA, 50 μg/ml heparin, 2.5 mM EDTA pH 8.0, 0.1% Tween® 20) for 1 h on a poly-l-ysine coated slide at 37 °C and hybridized in Hybmix with the specific probe over night at 37 °C. After hybridization, cells were washed with 2 × SSC and 1 × SSC at room temperature, each for 1 h and 0.5 × SSC at 37 °C and room temperature, each for 30 min. DNA was stained with Hoechst 33342 (Sigma). Microscopy studies were performed with a Leica AF6000 microscope and pictures were obtained by using the LEICA DFC360FX camera and the LAS AF 2.7.3.9 software (Leica). For deconvolution (Fig. 1E, 2A, 3F, Supplemantry Fig. S2A–C, Supplemantry Fig. S3D,E) z-stacks (10 stacks; 0.2 µm) were recorded and the maximal projection was deconvoluted with 3 iterations by the LAS AF 2.7.3.9 software (Leica).
Immunofluorescence experiments
The experiments were essentially carried out as described in24. For signal enhancement of GFP-tagged proteins, they were visualized using a primary anti-GFP and a secondary FITC-coupled antibody. Cells were grown to mid log phase (1 × 107 cells/ml) prior to a temperature shift to 16 °C for 1 h 15 min (Fig. 3G,H, Supplemantry Fig. S3F,G). Cells were fixed in a final concentration of 4% formaldehyde for 10 min at the restrictive temperature and a 50 min incubation at 25 °C. Cells were spheroblasted using zymolyase and placed on a poly-l-lysine coated slide. The cells were permeabilized with 0.1 M potassium phosphate buffer pH 6.5, 1.2 M sorbitol, 0.5% Triton® X-100. Afterwards they were blocked for 1 h in antibody blocking buffer (0.1 M Tris pH 9.0, 0.2 M NaCl, 5% heat-inactivated FCS and 0.3% tween). The buffer was removed and replaced with a 1:1.000 dilution of the primary anti-GFP (Thermo Fisher) mouse antibody in antibody blocking buffer and incubated for 2 h at room temperature. The cells were washed twice shortly with 1 × PBS, 0.5% tween and once for 30 min. Afterwards the cells were washed once with 1 × PBS and once with antibody blocking buffer each for 30 min. The secondary aqntibody (anti-mouse IgG-FITC; Sigma) was diluted 1:100 in antibody blocking buffer and incubated for 1 h at room temperature. Finally, the cells were washed once quickly with 1 × PBS, 0.5% tween and once for 30 min. Subsequently, cells were washed twice with 1 × PBS for 15 min each. DNA staining and microscopy was performed as described in the FISH experiment.
RNA co-immunoprecipitation experiments (RIP)
All yeast strains were grown to mid log phase (2 × 107 cells/ml). For RIP experiments seen in Fig. 1A,B,J,K, 2C,D and 6C,D (Supplemantry Fig. S1A,D, 2D, 6B) cells were cultured at 25 °C, in 1C, D, G-I; 2E-H; 4D-G and 6F (Supplemantry Fig. S1B,C, 2E, 4A) cells were shifted to a non-permissive temperature for 1 h 37 °C or 1 h and 15 min 16 °C respectively. Afterwards cells were harvested and lysed in RIP buffer (25 mM Tris HCl pH 7.5, 100 mM KCl, 0.2% (v/v) Triton X-100 (1% Triton X-100 for 2E), 0.2 mM PMSF, 5 mM DTT, 10 U RiboLock RNase Inhibitor (Thermo Scientific) and protease inhibitor (Roche) using the FastPrep®-24 machine (MP Biomedicals) three times for 30 s at 5.5 m/s. After centrifugation the supernatant was incubated for 1 h at 4 °C with GFP-Selector beads (NanoTag) (Fig. 1B,D,H,I,K, 2D,F–H, 4E,G and 6D). For TMG-cap-IPs total RNA was extracted from yeast lysates using trizol-chloroform (Ambion® RNA by Life technologies™). 50 µg of the total RNA was incubated for 1 h at 4 °C with 10 µl of the anti-2,2,7-trimethylguanosine-antibody (Calbiochem Milipore) coupled to sepharose beads (Fig. 6F).
The beads were washed five times with RIP buffer and for GFP-RIP split in two portions after the last washing step. Proteins were detected by western blot (1A, C, G and J; 2C, E; 4D, F and 6C). Eluates were purified via trizol-chloroform (Ambion® RNA by Life technologies™) extraction. The purified RNA was measured via Nanodrop and a defined amount of RNA was reverse transcribed with FastGene Scriptase II (Nippon Genetics) for subsequent qPCR analyses. All eluates were related in each strain to the lysates and substracted from the no-tag controls to account for strain-specific variations in the total amount. To normalize the total RNA amount, we used the mitochondrial 21S rRNA. In Figs. 2H and 6B, the enrichment of the unprocessed form was shown in comparison to total TLC1 amount.
Total RNA isolation
Total RNA Isolation was carried out with the NucleoSpin RNA Kit from Macherey–Nagel (Figs. 2I–K, 6E). All steps were performed according to the manufactures description, except step 7. The DNA digestion on the column was executed for 1 h. An additional DNA digestion step was carried out after elution of the RNA. For the additional DNA digest the eluted RNA was mixed with a 10th volume of the Reaction Buffer for rDNAse and 1 µl rDNAse, according to the manufactures description. The digest was incubated for 1 h at 37 °C, before it was terminated through sodium acetate ethanol precipitation. For this 0.1 volume 3 M sodium acetate, pH 5.2, 2.5 volumes of 99% pure ethanol and 1 µl glycoblue were mixed with the RNA and incubated over night at − 20 °C. The purified RNA was measured via Nanodrop and a defined amount of RNA was reverse transcribed with FastGene Scriptase II (Nippon Genetics) for subsequent qPCR analyses.
Nucleo-cytoplasmic fractionation experiment
For the detection of TLC1 in the cytoplasm (Fig. 6B, Supplementary Fig. S6A) cells were grown to mid log-phase (2 × 107 cells/ml). The cells were harvestet by centrifugation for 5 min at 4000 rpm. Cells were washed once with 1 ml YPD/ 1 M Sorbitol/ 2 mM DTT and resuspended in YPD/ 1 M Sorbitol/ 1 mM DTT. Cells were spheroblasted using 1 mg zymolyase (100 mg/ml) and after that diluted in 50 mL YPD/ 1 M Sorbitol for 30 min at 25 °C for recovery. Subsequently, the cells were shifted to 16 °C for 1 h 15 min (cse1-1 and cse1-1 mtr10∆). Cells were placed on ice, centrifuged at 2000 rpm for 10 min and the pelleted cells were resuspended in 500 μl Ficoll buffer (18% Ficoll 400, 10 mM HEPES pH 6.0) and 1 µl Ribolock. Cells were lysed by addition of 1 ml buffer A (50 mM NaCl, 1 mM MgCl2, 10 mM HEPES pH 6.0). The suspension was mixed and centrifuged at 4000 rpm for 15 min. The supernatant was used for cytoplasmic analyses. RNA was isolated using the Nucleo-Spin RNA Kit (Macherey and Nagel). The purified RNA was reverse transcribed with FastGene Scriptase II (Nippon Genetics) for subsequent qPCR analyses. All values were normalized to the amount of the mRNA present for RPL8A. To verify no nuclear contamination in the cytoplasmic fraction, aliquots of the samples were analyzed in Western blots for the presence of the cytoplasmic Zwf1 protein and the absence of the nuclear Nop1 protein (Fig. 6A).
GFP-microscopy
Cells were grown, treated and harvested as described in the FISH experiments. Cells were fixed with 3% formaldehyde for 2 min at room temperature, subsequently washed with 0.1 M phosphate buffer pH 6.5 and with P-solution (0.1 M phosphate buffer pH 6.5, 1.2 M Sorbitol), before an aliquot was added to a poly-l-lysine-coated slide for 30 min at 4 °C. Permeabilization of the cells, DNA staining and microscopy was performed as described in the FISH experiment.
Co-immunoprecipitation (IP) experiments
All yeast strains were grown to log phase (2–3 × 107 cells/ml). Afterwards, the cells were harvested and lysed in IP buffer (1 × PBS, 3 mM KCl, 2.5 mM MgCl2, 0,5% Triton X-100 and protease inhibitors from Roche). 35 µl of this lysate was loaded onto an SDS-gel (lysate lanes). The supernatant was incubated for 1 h at 4 °C with GFP-Selector beads (NanoTag) (Fig. 5A,B,D, Supplementary Fig. S5.1A, 5.2A, 5.3B) or with Myc-trap beads (Chromotek) (Fig. 5C, Supplementary Fig. S5.3A). The beads were washed five times with IP buffer, and finally resuspended in 35 µl SDS-sample buffer. The entire eluate sample was loaded onto the SDS-gels.
Subsequently, the proteins were detected by Western blot analyses with the indicated antibodies (GFP (Chromotek) 1:4,000; c-myc (9E10) (Santa Cruz) 1:1,000; Hem15 and Grx4 each 1:5,000 and Aco1 1:2,000 (U. Mühlenhoff); Nop1 (Santa Cruz) 1:4,000; Hdf1 (Yku70) (Santa Cruz) 1:4,000). Signals were detected with the Fusion SL system (PeqLab) and FusionFX7 Edge (Fusion FX Vilber). To be able to detect several proteins in one experiment, the western blots were cut horizontally according to the size of the desired proteins to be able to detect each stripe with individual antibodies.
Drop dilution analysis
Cells were grown to log phase (2–3 × 107 cells/ml) and diluted to 1 × 107 cells/ml. Tenfold serial dilutions to 1 × 103 cells/ml were prepared and 10 µl of each dilution was spotted onto full medium (YPD) agar plates that were subsequently incubated for 3 days at the indicated temperatures (Fig. 2B). Pictures were taken after 2 and 3 days with the Intelli Scan 1600 (Quanto technology) and the SilverFast Ai program.
Southern blot analysis
Yeast strains were either passaged in liquid culture (Fig. 3B,C, Supplementary Fig. S3A,B) or on solid agar plates (Fig. 3D, Supplementary Fig. S3C). For passaging in liquid culture, cells were inoculated with a starting concentration of 1 × 105 cells/ml. The strains were grown for 3 days per passage at 20 °C until they reached 1 × 108 cells/ml and were afterwards diluted again to 1 × 105. Overall 8 passages, corresponding to 80 generations, were carried out. Cell passages on solid agar plates were used for generating Type I survivors in tlc1∆ cells. The strains were freshly restreaked form the database and single colonies were restreaked after 2–3 days at 25 °C. One restreak comprises about 25 generations. Overall 5 restreaks were generated coresponding to about 125 generations.
Genomic DNA was isolated with the MasterPure Yeast DNA Purification Kit (Lucigen). This gDNA was digested with XhoI (Nippon Genetics) according to the manufactures description. 20 µg gDNA were loaded per lane and electrophoretically separated on a 1% TBE gel at 4 °C for 3 h. The gel was depurinated (250 mM HCl) for 15 min, denatured (1.5 M NaCl, 0.5 M NaOH) for 30 min, neutralized (1.5 M NaCl, 0.5 M Tris–HCL, pH 7.5) for 30 min and equilibrated in 20 × SSC (0.3 M Tri-sodium citrate, 3 M NaCl, pH 7.0) for 15 min. A capillary blot was used for the transfer onto a HybondN+ (GE Healthcare) membrane. After transfer, the membrane was crosslinked for 7 min by UV light (254 nm, 120,000 µJ/cm2) and then heated to 80 °C for 2 h. Subsequently, pre-hybridization was carried out for 1 h at 68 °C in pre-hybridization buffer (0.5 M sodium phosphate buffer pH 7,5; 7% (w/v) SDS; 1 mM EDTA). For hybridization, a digoxygenin-labeled probe, detecting the TG repeats of the telomeres, was added to the pre-hybridization solution and incubated over night at 37 °C with rotation. After washing, the southern blot was detected by using an anti-digoxygenin antibody coupled to alkaline phosphatase (Roche) and the labeling and detection starter kit (Roche).
Senescence assay
Yeast strains were passaged every 10 generations in fresh liquid medium (Fig. 3E). Strains were grown at the semi-permissive temperature of 20 °C.
Quantification
All experiments shown in this work were performed in biologically independent repetitions as indicated in the figure legends. Error bars represent the standard deviation. P values were calculated using a one or two-tailed, two-sample unequal variance t-test. P values are indicated as follows: ***p < 0.001, **p < 0.01, *p < 0.05.
Data availability
The datasets generated and/or analyzed for this study are available from the corresponding author upon reasonable request.
References
Armstrong, C. A. & Tomita, K. Fundamental mechanisms of telomerase action in yeasts and mammals: Understanding telomeres and telomerase in cancer cells. Open Biol. https://doi.org/10.1098/rsob.160338 (2017).
Vasianovich, Y. & Wellinger, R. J. Life and death of yeast telomerase RNA. J. Mol. Biol. https://doi.org/10.1016/j.jmb.2017.01.013 (2017).
Kupiec, M. Biology of telomeres: Lessons from budding yeast. FEMS Microbiol. Rev. 38, 144–171. https://doi.org/10.1111/1574-6976.12054 (2014).
Blackburn, E. H., Epel, E. S. & Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198. https://doi.org/10.1126/science.aab3389 (2015).
O’Sullivan, R. J. & Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. 11, 171–181. https://doi.org/10.1038/nrm2848 (2010).
Rousseau, P. & Autexier, C. Telomere biology: Rationale for diagnostics and therapeutics in cancer. RNA Biol. 12, 1078–1082. https://doi.org/10.1080/15476286.2015.1081329 (2015).
Gallardo, F. & Chartrand, P. Telomerase biogenesis: The long road before getting to the end. RNA Biol 5, 212–215 (2008).
Zappulla, D. C. Yeast telomerase RNA flexibly scaffolds protein subunits: Results and repercussions. Molecules https://doi.org/10.3390/molecules25122750 (2020).
Bosoy, D., Peng, Y., Mian, I. S. & Lue, N. F. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 278, 3882–3890. https://doi.org/10.1074/jbc.M210645200 (2003).
Rhee, H. S. & Pugh, B. F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301. https://doi.org/10.1038/nature10799 (2012).
Mouaikel, J., Verheggen, C., Bertrand, E., Tazi, J. & Bordonne, R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9, 891–901 (2002).
Seto, A. G., Zaug, A. J., Sobel, S. G., Wolin, S. L. & Cech, T. R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401, 177–180. https://doi.org/10.1038/43694 (1999).
Chapon, C., Cech, T. R. & Zaug, A. J. Polyadenylation of telomerase RNA in budding yeast. RNA 3, 1337–1351 (1997).
Jamonnak, N. et al. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17, 2011–2025. https://doi.org/10.1261/rna.2840711 (2011).
Noel, J. F., Larose, S., Abou Elela, S. & Wellinger, R. J. Budding yeast telomerase RNA transcription termination is dictated by the Nrd1/Nab3 non-coding RNA termination pathway. Nucleic Acids Res. 40, 5625–5636. https://doi.org/10.1093/nar/gks200 (2012).
Zappulla, D. C. & Cech, T. R. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. U.S.A. 101, 10024–10029. https://doi.org/10.1073/pnas.0403641101 (2004).
Coy, S., Volanakis, A., Shah, S. & Vasiljeva, L. The Sm complex is required for the processing of non-coding RNAs by the exosome. PLoS ONE 8, e65606. https://doi.org/10.1371/journal.pone.0065606 (2013).
Hass, E. P. & Zappulla, D. C. Repositioning the Sm-binding site in Saccharomyces cerevisiae telomerase RNA reveals RNP organizational flexibility and Sm-Directed 3’-end formation. Non-coding RNA https://doi.org/10.3390/ncrna6010009 (2020).
Fisher, T. S., Taggart, A. K. & Zakian, V. A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 11, 1198–1205. https://doi.org/10.1038/nsmb854 (2004).
Hass, E. P. & Zappulla, D. C. The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. Elife https://doi.org/10.7554/eLife.07750 (2015).
Garcia, P. D. et al. Stability and nuclear localization of yeast telomerase depend on protein components of RNase P/MRP. Nat. Commun. 11, 2173. https://doi.org/10.1038/s41467-020-15875-9 (2020).
Laterreur, N. et al. The yeast telomerase module for telomere recruitment requires a specific RNA architecture. RNA 24, 1067–1079. https://doi.org/10.1261/rna.066696.118 (2018).
Lemieux, B. et al. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell 165, 1171–1181. https://doi.org/10.1016/j.cell.2016.04.018 (2016).
Wu, H., Becker, D. & Krebber, H. Telomerase RNA TLC1 shuttling to the cytoplasm requires mRNA export factors and is important for telomere maintenance. Cell Rep. 8, 1630–1638. https://doi.org/10.1016/j.celrep.2014.08.021 (2014).
Becker, D. et al. Nuclear pre-snRNA export is an essential quality assurance mechanism for functional spliceosomes. Cell Rep. 27, 3199–3214. https://doi.org/10.1016/j.celrep.2019.05.031 (2019).
Soheilypour, M. & Mofrad, M. R. K. Quality control of mRNAs at the entry of the nuclear pore: Cooperation in a complex molecular system. Nucleus 9, 202–211. https://doi.org/10.1080/19491034.2018.1439304 (2018).
Soheilypour, M. & Mofrad, M. R. K. Regulation of the affinity between RNA-binding proteins and the export receptor enables nuclear basket proteins to distinguish and retain aberrant mRNAs. Sci. Rep. https://doi.org/10.1038/srep35380 (2016).
Zander, G. & Krebber, H. Quick or quality? How mRNA escapes nuclear quality control during stress. RNA Biol. 14, 1642–1648. https://doi.org/10.1080/15476286.2017.1345835 (2017).
Gallardo, F., Olivier, C., Dandjinou, A. T., Wellinger, R. J. & Chartrand, P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 27, 748–757. https://doi.org/10.1038/emboj.2008.21 (2008).
Vasianovich, Y., Bajon, E. & Wellinger, R. J. Telomerase biogenesis requires a novel Mex67 function and a cytoplasmic association with the Sm(7) complex. Elife https://doi.org/10.7554/eLife.60000 (2020).
Ferrezuelo, F., Steiner, B., Aldea, M. & Futcher, B. Biogenesis of yeast telomerase depends on the importin mtr10. Mol. Cell. Biol. 22, 6046–6055 (2002).
Bordonne, R. Functional characterization of nuclear localization signals in yeast Sm proteins. Mol. Cell. Biol. 20, 7943–7954 (2000).
Hood, J. K. & Silver, P. A. Cse1p is required for export of Srp1p/importin-alpha from the nucleus in Saccharomyces cerevisiae. J. Biol. Chem. 273, 35142–35146 (1998).
Lange, A., Fasken, M. B., Stewart, M. & Corbett, A. H. Dissecting the roles of Cse1 and Nup2 in classical NLS-cargo release in vivo. Traffic 21, 622–635. https://doi.org/10.1111/tra.12759 (2020).
Bischoff, F. R. & Gorlich, D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 419, 249–254 (1997).
Bischoff, F. R., Klebe, C., Kretschmer, J., Wittinghofer, A. & Ponstingl, H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. U.S.A. 91, 2587–2591 (1994).
Dasso, M. The Ran GTPase: Theme and variations. Curr Biol 12, R502-508 (2002).
Bajon, E., Laterreur, N. & Wellinger, R. J. A single templating RNA in yeast telomerase. Cell Rep. 12, 441–448. https://doi.org/10.1016/j.celrep.2015.06.045 (2015).
Mozdy, A. D. & Cech, T. R. Low abundance of telomerase in yeast: Implications for telomerase haploinsufficiency. RNA 12, 1721–1737. https://doi.org/10.1261/rna.134706 (2006).
Hu, Y. et al. Telomerase-null survivor screening identifies novel telomere recombination regulators. PLoS Genet. 9, e1003208. https://doi.org/10.1371/journal.pgen.1003208 (2013).
Lundblad, V. & Blackburn, E. H. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73, 347–360. https://doi.org/10.1016/0092-8674(93)90234-h (1993).
Teng, S. C. & Zakian, V. A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 8083–8093. https://doi.org/10.1128/mcb.19.12.8083 (1999).
Grandin, N. & Charbonneau, M. Control of the yeast telomeric senescence survival pathways of recombination by the Mec1 and Mec3 DNA damage sensors and RPA. Nucleic Acids Res. 35, 822–838. https://doi.org/10.1093/nar/gkl1081 (2007).
Hawkins, C. & Friedman, K. L. Normal telomere length maintenance in Saccharomyces cerevisiae requires nuclear import of the ever shorter telomeres 1 (Est1) protein via the importin alpha pathway. Eukaryot. Cell 13, 1036–1050. https://doi.org/10.1128/ec.00115-14 (2014).
Gill, T., Aulds, J. & Schmitt, M. E. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J. Cell Biol. 173, 35–45. https://doi.org/10.1083/jcb.200512025 (2006).
Perederina, A., Esakova, O., Quan, C., Khanova, E. & Krasilnikov, A. S. Eukaryotic ribonucleases P/MRP: The crystal structure of the P3 domain. EMBO J. 29, 761–769. https://doi.org/10.1038/emboj.2009.396 (2010).
Chen, H. et al. Structural insights into yeast telomerase recruitment to telomeres. Cell 172, 331–343. https://doi.org/10.1016/j.cell.2017.12.008 (2018).
Matera, A. G. & Wang, Z. A day in the life of the spliceosome. Nat. Rev. 15, 108–121. https://doi.org/10.1038/nrm3742 (2014).
Lei, E. P., Krebber, H. & Silver, P. A. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15, 1771–1782 (2001).
Zander, G. et al. mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature 540, 593–596. https://doi.org/10.1038/nature20572 (2016).
Mattaj, I. W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46, 905–911 (1986).
Lund, M. K. & Guthrie, C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell 20, 645–651 (2005).
Tieg, B. & Krebber, H. Dbp5: From nuclear export to translation. Biochem. Biophys. Acta. 1829, 791–798. https://doi.org/10.1016/j.bbagrm.2012.10.010 (2013).
Lendvay, T. S., Morris, D. K., Sah, J., Balasubramanian, B. & Lundblad, V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144, 1399–1412 (1996).
Lundblad, V. & Szostak, J. W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57, 633–643 (1989).
Acknowledgements
We are grateful to R. Bordonné, E. Hurt, U. Mühlenhoff, R. Lill, V. Lundblad, E. O’Shea, P.A. Silver, A. Tartakoff and K. Weis for providing plasmids, strains or antibodies. This work was supported from the Deutsche Forschungsgemeinschaft (DFG) (KR1779/9-1), the SFB860 (to H.K.) and the University of Göttingen (to H.K).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Experiments were designed and data interpreted by A.G.H., D.B., J-P.L. and H.K.; all experiments were performed by A.G.H. except 1 E, which was done by D.B; 2A by A.G.H. and D.B.; 2B, 4A,B,C by A.G.H and J.-P.L.. The manuscript was written by H.K.; all authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hirsch, A.G., Becker, D., Lamping, JP. et al. Unraveling the stepwise maturation of the yeast telomerase including a Cse1 and Mtr10 mediated quality control checkpoint. Sci Rep 11, 22174 (2021). https://doi.org/10.1038/s41598-021-01599-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01599-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.