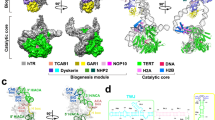
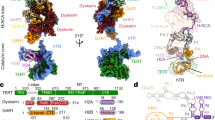
Abstract
Telomerase is a special reverse transcriptase ribonucleoprotein dedicated to the synthesis of telomere repeats that protect chromosome ends. Among reverse transcriptases, telomerase is unique in using a stably associated RNA with an embedded template to synthesize a specified sequence. Moreover, it is capable of iteratively copying the same template region (repeat addition processivity) through multiple rounds of RNA–DNA unpairing and reannealing, that is, the translocation reaction. Biochemical analyses of telomerase over the past 3 decades in protozoa, fungi and mammals have identified structural elements that underpin telomerase mechanisms and have led to models that account for the special attributes of telomerase. Notably, these findings and models can now be interpreted and adjudicated through recent cryo-EM structures of Tetrahymena and human telomerase holoenzyme complexes in association with substrates and regulatory proteins. Collectively, these structures reveal the intricate protein–nucleic acid interactions that potentiate telomerase’s unique translocation reaction and clarify how this enzyme reconfigures the basic reverse transcriptase scaffold to craft a polymerase dedicated to the synthesis of telomere DNA. Among the many new insights is the resolution of the telomerase ‘anchor site’ proposed more than 3 decades ago. The structures also highlight the nearly universal conservation of a protein–protein interface between an oligonucleotide/oligosaccharide-binding (OB)-fold regulatory protein and the telomerase catalytic subunit, which enables spatial and temporal regulation of telomerase function in vivo. In this Review, we discuss key features of the structures in combination with relevant functional analyses. We also examine conserved and divergent aspects of telomerase mechanisms as gleaned from studies in different model organisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data generated and/or analyzed during the current study are included within the paper and its Supplementary Information or are available from the corresponding author upon reasonable request.
Code availability
The ColabFold modeling software is available at GitHub.
References
de Lange, T. A loopy view of telomere evolution. Front. Genet. 6, 321 (2015).
Garavis, M., Gonzalez, C. & Villasante, A. On the origin of the eukaryotic chromosome: the role of noncanonical DNA structures in telomere evolution. Genome Biol. Evol. 5, 1142–1150 (2013).
Blackburn, E. H., Epel, E. S. & Lin, J. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198 (2015).
Blasco, M. A. Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 6, 611–622 (2005).
Jafri, M. A., Ansari, S. A., Alqahtani, M. H. & Shay, J. W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 8, 69 (2016).
Armanios, M. The role of telomeres in human disease. Annu. Rev. Genomics Hum. Genet. 23, 363–381 (2022).
Bonnell, E., Pasquier, E. & Wellinger, R. J. Telomere replication: solving multiple end replication problems. Front. Cell Dev. Biol. 9, 668171 (2021).
Lue, N. F. Evolving linear chromosomes and telomeres: a C-strand-centric view. Trends Biochem. Sci. 43, 314–326 (2018).
Lim, C. J. & Cech, T. R. Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat. Rev. Mol. Cell Biol. 22, 283–298 (2021).
Wu, R. A., Upton, H. E., Vogan, J. M. & Collins, K. Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem. 86, 439–460 (2017).
Autexier, C. & Lue, N. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75, 493–517 (2006).
Liu, B. et al. Structure of active human telomerase with telomere shelterin protein TPP1. Nature 604, 578–583 (2022). Report of cryo-EM structures (3.5-Å and 3.7-Å resolutions) of DNA-bound human telomerase holoenzyme in complex with or without TPP1 that highlight the three-way TPP1–TEN–IFDTRAP interface and reveal a 4-bp RNA–DNA duplex in the catalytic center.
Nguyen, T. H. D. et al. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557, 190–195 (2018). Report of cryo-EM structures (8.2-Å and 7.7-Å resolutions) of substrate-bound human telomerase holoenzyme showing two RNA-tethered lobes consisting of a catalytic core and a biogenesis module.
Schmidt, J. C. & Cech, T. R. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 29, 1095–1105 (2015).
MacNeil, D. E., Bensoussan, H. J. & Autexier, C. Telomerase regulation from beginning to the end. Genes 7, 64 (2016).
Greider, C. W. & Blackburn, E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413 (1985).
Morin, G. B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59, 521–529 (1989).
Lingner, J. & Cech, T. R. Purification of telomerase from Euplotes aediculatus: requirement of a 3′ overhang. Proc. Natl Acad. Sci. USA 93, 10712–10717 (1996).
Nakamura, T. M. et al. Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955–959 (1997).
Collins, K. & Gandhi, L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA 95, 8485–8490 (1998).
Nakamura, T. M. & Cech, T. R. Reversing time: origin of telomerase. Cell 92, 587–590 (1998).
Fitzgerald, M. S. et al. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA 96, 14813–14818 (1999).
Malik, H. S., Burke, W. D. & Eickbush, T. H. Putative telomerase catalytic subunits from Giardia lamblia and Caenorhabditis elegans. Gene 251, 101–108 (2000).
Singer, M. S. & Gottschling, D. E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266, 404–409 (1994).
Blasco, M. A. et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34 (1997).
Harley, C. B., Futcher, A. B. & Greider, C. W. Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 (1990).
de Lange, T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 52, 223–247 (2018).
Kim, N. W. et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994).
Wright, W. E., Piatyszek, M. A., Rainey, W. E., Byrd, W. & Shay, J. W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179 (1996).
Mitchell, J. R., Wood, E. & Collins, K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 (1999).
Greider, C. W. Telomerase is processive. Mol. Cell. Biol. 11, 4572–4580 (1991).
Autexier, C. & Greider, C. W. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 15, 2227–2239 (1995).
Lai, C. K., Miller, M. C. & Collins, K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 16, 415–420 (2002).
Tzfati, Y., Fulton, T. B., Roy, J. & Blackburn, E. H. Template boundary in a yeast telomerase specified by RNA structure. Science 288, 863–867 (2000).
Podlevsky, J. D. & Chen, J. J. Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 13, 720–732 (2016).
Brown, A. F. et al. A self-regulating template in human telomerase. Proc. Natl Acad. Sci. USA 111, 11311–11316 (2014).
Jacobs, S. A., Podell, E. R. & Cech, T. R. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 13, 218–225 (2006).
Romi, E. et al. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc. Natl Acad. Sci. USA 104, 8791–8796 (2007).
Collins, K. & Greider, C. W. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 7, 1364–1376 (1993).
Harrington, L. A. & Greider, C. W. Telomerase primer specificity and chromosome healing. Nature 353, 451–454 (1991).
Lee, M. S. & Blackburn, E. H. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol. 13, 6586–6599 (1993).
Morin, G. B. Recognition of a chromosome truncation site associated with α-thalassaemia by human telomerase. Nature 353, 454–456 (1991).
Lue, N. F., Lin, Y. C. & Mian, I. S. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol. Cell. Biol. 23, 8440–8449 (2003).
Xie, M., Podlevsky, J. D., Qi, X., Bley, C. J. & Chen, J. J. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 38, 1982–1996 (2010).
Jiang, J. et al. Structure of telomerase with telomeric DNA. Cell 173, 1179–1190 (2018). Report of a 4.8-Å-resolution cryo-EM structure of T. thermophila telomerase bound to telomeric DNA, revealing an independent structural module composed of the TEN domain and IFDTRAP, which forms a partial lid over the TERT ring; this unique ring-plus-lid conformation of TERT provides the structural underpinning for telomerase to accomplish processive telomere DNA synthesis.
Wang, Y., Gallagher-Jones, M., Susac, L., Song, H. & Feigon, J. A structurally conserved human and Tetrahymena telomerase catalytic core. Proc. Natl Acad. Sci. USA 117, 31078–31087 (2020). Using multiple sequence alignments and statistical coupling analysis on all identified TERTs, this study finds that the two β-strands implicated in the TEN–IFDTRAP interaction represent two of the most well-conserved, TERT-specific sequence motifs that can serve as a signature for archetypal telomerases.
Hong, K. et al. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol. Cell. Biol. 33, 3962–3971 (2013).
Armstrong, C. A., Pearson, S. R., Amelina, H., Moiseeva, V. & Tomita, K. Telomerase activation after recruitment in fission yeast. Curr. Biol. 24, 2006–2011 (2014).
Talley, J. M., DeZwaan, D. C., Maness, L. D., Freeman, B. C. & Friedman, K. L. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J. Biol. Chem. 286, 26431–26439 (2011).
Wang, F. et al. The Pot1–TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510 (2007).
Gillis, A. J., Schuller, A. P. & Skordalakes, E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455, 633–637 (2008).
Cohn, M. & Blackburn, E. H. Telomerase in yeast. Science 269, 396–400 (1995).
Sekne, Z., Ghanim, G. E., van Roon, A. M. & Nguyen, T. H. D. Structural basis of human telomerase recruitment by TPP1–POT1. Science 375, 1173–1176 (2022). Report of cryo-EM structures (3.2-Å and 3.9-Å resolutions) of DNA-bound human telomerase holoenzyme in complex with TPP1 or TPP1–POT1 that highlight the three-way TPP1–TEN–IFDTRAP interface and a path for telomeric DNA from POT1 to the catalytic center with contacts to TEN, IFD, motif 3 and the CTE.
Ghanim, G. E. et al. Structure of human telomerase holoenzyme with bound telomeric DNA. Nature 593, 449–453 (2021). Report of cryo-EM structures (3.8-Å and 3.4-Å resolutions) of human telomerase holoenzyme bound to telomeric DNA revealing the presence of a histone H2A–H2B dimer in the catalytic core and a detailed structure of the dyskerin–dyskerin interface.
He, Y. et al. Structures of telomerase at several steps of telomere repeat synthesis. Nature 593, 454–459 (2021). Cryo-EM structures (3.3-Å, 4.4-Å and 3.8-Å resolutions) of T. thermophila telomerase with telomeric DNA at different steps of nucleotide addition, providing evidence for the maintenance of a short 4-bp RNA–DNA duplex during successive nucleotide addition and supporting a role of the catalytic core in anchor site function.
Wan, F. et al. Zipper head mechanism of telomere synthesis by human telomerase. Cell Res. 31, 1275–1290 (2021). Characterization of a 3.54-Å-resolution structure of the catalytic module of human telomerase with a 24-nucleotide DNA species in combination with molecular dynamic simulation, which revealed the role of Leu980 in the thumb helix in acting as a ‘zipper head’ to limit the RNA–DNA duplex at the active site to just three canonical base pairs.
He, Y. & Feigon, J. Telomerase structural biology comes of age. Curr. Opin. Struct. Biol. 76, 102446 (2022).
Nguyen, T. H. D. Structural biology of human telomerase: progress and prospects. Biochem. Soc. Trans. 49, 1927–1939 (2021).
Smith, E. M., Pendlebury, D. F. & Nandakumar, J. Structural biology of telomeres and telomerase. Cell. Mol. Life Sci. 77, 61–79 (2020).
Lue, N. F. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem. Sci. 35, 8–17 (2010).
Grill, S., Tesmer, V. M. & Nandakumar, J. The N terminus of the OB domain of telomere protein TPP1 is critical for telomerase action. Cell Rep. 22, 1132–1140 (2018).
Hu, X., Liu, J., Jun, H. I., Kim, J. K. & Qiao, F. Multi-step coordination of telomerase recruitment in fission yeast through two coupled telomere–telomerase interfaces. eLife 5, e15470 (2016).
Nandakumar, J. et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289 (2012).
Sexton, A. N., Youmans, D. T. & Collins, K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J. Biol. Chem. 287, 34455–34464 (2012).
Zaug, A. J., Podell, E. R., Nandakumar, J. & Cech, T. R. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 24, 613–622 (2010).
Zhong, F. L. et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 (2012).
Jiang, J. et al. The architecture of Tetrahymena telomerase holoenzyme. Nature 496, 187–192 (2013). Report of a cryo-EM structure (25-Å resolution) of a telomerase holoenzyme revealing an extensive network of protein associations, the interaction of p50 with the catalytic core of T. thermophila telomerase and the identification of p50 as functionally equivalent to human TPP1 in regulating enzyme processivity.
Lee, J., Mandell, E. K., Tucey, T. M., Morris, D. K. & Lundblad, V. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat. Struct. Mol. Biol. 15, 990–997 (2008).
Yu, E. Y., Wang, F., Lei, M. & Lue, N. F. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat. Struct. Mol. Biol. 15, 985–989 (2008).
Schmidt, J. C., Dalby, A. B. & Cech, T. R. Identification of human TERT elements necessary for telomerase recruitment to telomeres. eLife 3, e03563 (2014).
Min, B. & Collins, K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell 36, 609–619 (2009).
Jiang, J. et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350, aab4070 (2015). Report of cryo-EM structures (9.4-Å and 8.9-Å resolutions) of the Tetrahymena telomerase holoenzyme, revealing the N-terminal, functional domain of p50 to be an OB fold juxtaposed to the TERT protein and presenting evidence that IFD independently contacts p50 and the TEN domain.
Chu, T. W., D’Souza, Y. & Autexier, C. The insertion in fingers domain in human telomerase can mediate enzyme processivity and telomerase recruitment to telomeres in a TPP1-dependent manner. Mol. Cell. Biol. 36, 210–222 (2016).
Tesmer, V. M., Smith, E. M., Danciu, O., Padmanaban, S. & Nandakumar, J. Combining conservation and species-specific differences to determine how human telomerase binds telomeres. Proc. Natl Acad. Sci. USA 116, 26505–26515 (2019).
D’Souza, Y., Chu, T. W. & Autexier, C. A translocation-defective telomerase with low levels of activity and processivity stabilizes short telomeres and confers immortalization. Mol. Biol. Cell 24, 1469–1479 (2013).
Lue, N. F. & Li, Z. Modeling and structure function analysis of the putative anchor site of yeast telomerase. Nucleic Acids Res. 35, 5213–5222 (2007).
Wyatt, H. D., Tsang, A. R., Lobb, D. A. & Beattie, T. L. Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS ONE 4, e7176 (2009).
Meier, B. et al. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PLoS Genet. 2, e18 (2006).
Osanai, M., Kojima, K. K., Futahashi, R., Yaguchi, S. & Fujiwara, H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle). Gene 376, 281–289 (2006).
Akiyama, B. M., Parks, J. W. & Stone, M. D. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA–DNA hybrids. Nucleic Acids Res. 43, 5537–5549 (2015).
Lue, N. F. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 280, 26586–26591 (2005).
Robart, A. R. & Collins, K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol. Cell 42, 308–318 (2011).
Patrick, E. M., Slivka, J. D., Payne, B., Comstock, M. J. & Schmidt, J. C. Observation of processive telomerase catalysis using high-resolution optical tweezers. Nat. Chem. Biol. 16, 801–809 (2020).
Jansson, L. I. et al. Telomere DNA G-quadruplex folding within actively extending human telomerase. Proc. Natl Acad. Sci. USA 116, 9350–9359 (2019).
Finger, S. N. & Bryan, T. M. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 36, 1260–1272 (2008).
Sealey, D. C. et al. The N-terminus of hTERT contains a DNA-binding domain and is required for telomerase activity and cellular immortalization. Nucleic Acids Res. 38, 2019–2035 (2010).
Shastry, S., Steinberg-Neifach, O., Lue, N. & Stone, M. D. Direct observation of nucleic acid binding dynamics by the telomerase essential N-terminal domain. Nucleic Acids Res. 46, 3088–3102 (2018).
Xia, J., Peng, Y., Mian, I. S. & Lue, N. F. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20, 5196–5207 (2000).
Yen, W. F., Chico, L., Lei, M. & Lue, N. F. Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc. Natl Acad. Sci. USA 108, 20370–20375 (2011).
Wu, R. A., Tam, J. & Collins, K. DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J. 36, 1908–1927 (2017).
Upton, H. E., Hong, K. & Collins, K. Direct single-stranded DNA binding by Teb1 mediates the recruitment of Tetrahymena thermophila telomerase to telomeres. Mol. Cell. Biol. 34, 4200–4212 (2014).
Zeng, Z. et al. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc. Natl Acad. Sci. USA 108, 20357–20361 (2011).
Petrova, O. A. et al. Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase. Nucleic Acids Res. 46, 1525–1540 (2018).
Zhai, L. T. et al. Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi. Nucleic Acids Res. 49, 4768–4781 (2021).
Wu, R. A. & Collins, K. Human telomerase specialization for repeat synthesis by unique handling of primer–template duplex. EMBO J. 33, 921–935 (2014).
Huard, S., Moriarty, T. J. & Autexier, C. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 31, 4059–4070 (2003).
Qi, X. et al. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. EMBO J. 31, 150–161 (2011).
Nowak, E. et al. Ty3 reverse transcriptase complexed with an RNA–DNA hybrid shows structural and functional asymmetry. Nat. Struct. Mol. Biol. 21, 389–396 (2014).
Yang, W. & Lee, Y. S. A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat. Struct. Mol. Biol. 22, 844–847 (2015).
Lue, N. F. & Peng, Y. Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res. 25, 4331–4337 (1997).
Soudet, J., Jolivet, P. & Teixeira, M. T. Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol. Cell 53, 954–964 (2014).
Gunisova, S. et al. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15, 546–559 (2009).
Forstemann, K. & Lingner, J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 6, 361–366 (2005).
Zhang, Y. et al. Phosphorylation of TPP1 regulates cell cycle-dependent telomerase recruitment. Proc. Natl Acad. Sci. USA 110, 5457–5462 (2013).
Taggart, A. K., Teng, S. C. & Zakian, V. A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297, 1023–1026 (2002).
Wu, Y. & Zakian, V. A. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc. Natl Acad. Sci. USA 108, 20362–20369 (2011).
Tucey, T. M. & Lundblad, V. Regulated assembly and disassembly of the yeast telomerase quaternary complex. Genes Dev. 28, 2077–2089 (2014).
Hsu, M., Yu, E. Y., Singh, S. M. & Lue, N. F. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot. Cell 6, 1330–1338 (2007).
Pennock, E., Buckley, K. & Lundblad, V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104, 387–396 (2001).
Padmanaban, S., Tesmer, V. M. & Nandakumar, J. Interaction hub critical for telomerase recruitment and primer–template handling for catalysis. Life Sci. Alliance 6, e202201727 (2023).
Sandhu, R., Sharma, M., Wei, D. & Xu, L. The structurally conserved TELR region on shelterin protein TPP1 is essential for telomerase processivity but not recruitment. Proc. Natl Acad. Sci. USA 118, e2024889118 (2021).
Rao, T. et al. Structure of Est3 reveals a bimodal surface with differential roles in telomere replication. Proc. Natl Acad. Sci. USA 111, 214–218 (2014).
Frank, A. K. et al. The shelterin TIN2 subunit mediates recruitment of telomerase to telomeres. PLoS Genet. 11, e1005410 (2015).
Pike, A. M., Strong, M. A., Ouyang, J. P. T. & Greider, C. W. TIN2 functions with TPP1/POT1 to stimulate telomerase processivity. Mol. Cell. Biol. 39, e00593-18 (2019).
Savage, S. A. et al. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 82, 501–509 (2008).
Zaug, A. J., Goodrich, K. J., Song, J. J., Sullivan, A. E. & Cech, T. R. Reconstitution of a telomeric replicon organized by CST. Nature 608, 819–825 (2022).
Cai, S. W. et al. Cryo-EM structure of the human CST-Polα/primase complex in a recruitment state. Nat. Struct. Mol. Biol. 29, 813–819 (2022).
He, Q. et al. Structures of the human CST-Polα–primase complex bound to telomere templates. Nature 608, 826–832 (2022).
He, Y. et al. Structure of Tetrahymena telomerase-bound CST with polymerase α–primase. Nature 608, 813–818 (2022).
Acknowledgements
We apologize to our colleagues whose works are not cited due to space constraints. We thank J. Feigon and K. Nguyen for insightful comments on the manuscript, and we thank A. Young for discussion and helpful suggestions. Work in our laboratories is supported by the Canadian Institutes of Health Research (grant PJT-166130) and a Natural Sciences and Engineering Research Council of Canada Discovery grant (C.A.) and by NSF MCB-1817331, NIH GM107287 and Hearst Endowed Faculty Fellow Fund (N.F.L.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Sara Osman, Carolina Perdigoto and Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lue, N.F., Autexier, C. Orchestrating nucleic acid–protein interactions at chromosome ends: telomerase mechanisms come into focus. Nat Struct Mol Biol 30, 878–890 (2023). https://doi.org/10.1038/s41594-023-01022-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-01022-7