Abstract
Defence systems against microbial pathogens are present in most living beings. The German cockroach Blattella germanica requires these systems to adapt to unhealthy environments with abundance of pathogenic microbes, in addition to potentially control its symbiotic systems. To handle this situation, four antimicrobial gene families (defensins, termicins, drosomycins and attacins) were expanded in its genome. Remarkably, a new gene family (blattellicins) emerged recently after duplication and fast evolution of an attacin gene, which is now encoding larger proteins with the presence of a long stretch of glutamines and glutamic acids. Phylogenetic reconstruction, within Blattellinae, suggests that this duplication took place before the divergence of Blattella and Episymploce genera. The latter harbours a long attacin gene (pre-blattellicin), but the absence of the encoded Glx-region suggests that this element evolved recently in the Blattella lineage. A screening of AMP gene expression in available transcriptomic SR projects of B. germanica showed that, while some AMPs are expressed during almost the whole development, others are restricted to shorter periods. Blattellicins are highly expressed only in adult females. None of the available SR tissue projects could be associated with blattellicins’ expression, suggesting that it takes place in other tissues, maybe the gut.
Similar content being viewed by others
Introduction
Multicellular eukaryotic genomes encode large sets of genes involved in defence mechanisms against microbial pathogens. The innate immune system controls the production of a variety of different types of proteins named antimicrobial peptides (AMPs), found in both vertebrate and invertebrate species. In insects, after the description of the first AMP in Hyalophora cecropia 1, hundreds were reported and are available in protein databases2,3. The German cockroach Blattella germanica (Order Blattodea) is a global pest in human settings including hospitals and schools. It is a vector of disease agents due to its lifestyle in unhealthy habitats. To defend against microbial pathogens, B. germanica, as other cockroaches, contains genes encoding AMPs. The sequencing of the cockroach genomes of Periplaneta americana4 and B. germanica5,6 revealed the presence of at least several types of AMP genes, encoding Attacins, Defensins, Drosomycins, Pro-rich peptides and Termicins.
AMPs are small proteins, usually below 100 residues3. Most are generally cationic and amphipathic, but there are more than 100 examples of naturally occurring anionic peptides with antimicrobial potential7. This implies that electrostatic interactions alone cannot explain their mode of action8. AMPs and lysozymes are among the main effector molecules that insects use to defend against those pathogens that arrive to the midgut or get inside the body. They are included among the type of proteins that induce pathogen death via lysis9. However, in contrast to lysozymes, AMPs synthesis used to be induced by several mechanisms such as the Toll and Imd signalling pathways. In insects, they are produced at diverse epithelial surfaces or at internal tissues such as fat body cells, to be released to the haemolymph and distributed to the whole body. The classically considered mode of action was their binding to microbial membranes, followed by the production of pores, membrane permeabilization and cell lysis. However, diverse types of actions on specific targets such as inhibition of protein synthesis or of bacterial cytokinesis have also been reported10,11.
The characterization of genes repertoires in plant and animal genomes revealed that most organisms harbour from five to ten distinct AMP gene families, frequently composed by several paralogous genes11. The evolutionary histories of AMP gene families during insect evolution were very dynamic with frequent duplications, deletions and pseudogenization events, but also de novo gene formation11,12. Initially, AMP proteins were considered to be not specific of concrete microbial species but to affect a broad range of organisms (Gram-negative bacteria, Gram-positive bacteria, fungi, etc.). However, recent studies based on Drosophila AMP gene inactivation revealed a complex pattern with some AMP gene products acting on reduced number of species, while others having a taxonomically broader effect10.
One additional function of AMPs is the regulation of endosymbiont or exosymbiont communities. In the weevil genus Sitophilus, one specific AMP, Coleoptericin A, evolved to control the mutualist endosymbiont Candidatus Sodalis pierantonius13 by preventing its escape from the bacteriocytes and its spread to other cells14. Permeabilization of symbiont membranes to solve the problem of the transport of metabolites between host cells and endosymbionts through sublethal AMP doses was also suggested15. B. germanica harbours specialized cells in the fat body, named bacteriocytes, which contain thousands of cells of the bacterial endosymbiont Blattabacterium cuenoti (hereafter, Blattabacterium). This symbiotic relationship is at least 150 My old and, with the exception of some endosymbiont losses, Blattabacterium and Blattodea hosts have been co-evolving since then16,17,18,19. The main role of the endosymbiont is to participate in host’s nitrogen economy20,21. Additionally, cockroaches are unique among insects as they also harbour a complex and rich microbiota in the hindgut, which composition and structure could be determined by the effect of specific AMPs22,23,24.
Antimicrobial peptides may serve as an alternative to solve the problem of antibiotic resistance in bacteria and have many biotechnological applications. In this work, we have carried out a thorough characterization of B. germanica AMPs describing 39 genes including an unusual Glx-rich new attacin-derived type that we have called blattellicin. The expression of each AMP gene may occur during the whole development or may be restricted to some stages, as it is the case of blattellicins.
Results
Genes encoding proteins with AMP domains in the genome of B. germanica
To identify annotated genes with AMP functions in the genome of B. germanica6 two strategies were used. The first was the search for product names including the terms defense, drosomycin, tenecin, phormicin, attacin and coleoptericin. The second was the search of annotated Pfam domains related to antimicrobial peptides. They are included in three clan domains of Pfam database: Knottin_1 (CL0054, Scorpion toxin-like knottin superfamily), Defensin (CL0075, Defensin/myotoxin-like superfamily) and Omega_toxin (CL0083, Omega toxin-like). The five detected Pfam domains were: PF11581 (Argos), PF03769 (Attacin_C), PF01097 (Defensin_2), PF00304 (Gamma-thionin) and PF11415 (Toxin_37). After the removal of C0J52_07645 (Giant-lens protein) and C0J52_08617 (putative defense protein 3), because they do not encode AMPs, 24 coding genes were retained (Supplementary Table 1). They were initially classified into the following groups: (i) Defensin_2 proteins (hereafter Defensin) (10 CDS, including two with the annotation partial = 5′), (ii) Drosomycin (Gamma-thionin domain) (10 CDS), (iii) Termicin (Toxin_37 domain) (3 CDS) and (iv) the CDS C0J52_26498. The latter, annotated as hypothetical protein, was a long protein (541 amino acids) with an Attacin_C domain. However, a less stringent domain analysis showed the potential presence of two or three additional domains in this protein with similarities with Attacin_C and Coleoptericin (PF06286).
In order to revise the annotated AMP coding genes, several B. germanica RNA-Seq SRA experiments (PRJNA389591) were screened for their expression using BLASTN and several AMP CDS as queries. Among the SRA runs with abundance of AMP reads, the RNA-Seq run SRR6784710 (whole body, adult female) was selected. Run SRR6784710 was assembled with de novo Trinity25 and a transcript database was created.
The annotated genome was compared with the transcript database with the aim of identifying the complete sets of AMP genes for each class. After careful revision, we identified 39 AMP genes (belonging to five types: defensins, termicins, drosomycins, attacins-like and blattellicins), which will be described below. Thirty-four of them were distributed in ten genome scaffolds and five genes were unplaced (Table 1; Supplementary Table 2).
Defensin AMP genes
Ten annotated AMP CDS with a Defensin domain were used as queries against the SRR6784710 transcript database with BLASTN (e-value = 1.0E−20). All of them produced hits with at least one transcript. In total, 16 different transcripts were identified. The transcript abundance ranged from TPM (transcripts per million transcripts) values of 323.64–0.00.
Information on genome annotation and the assembled transcripts was compared (see Materials & Methods) identifying 16 defensin genes (Supplementary Tables 2 and 3). They received the names of defensin_g1 to defensin_g16, with defensin_g1 and defensin_g16 including two alternative isoforms that do not affect the coding region. Defensin_g1 isoforms i1 and i2 differed in the removal or not of a 3′-UTR intron, whereas the two isoforms of defensin_g16 differed in the use of different poly(A) signals.
Defensin genes (except defensin_g1 that was unplaced) were clustered in four scaffolds. The unplaced defensin_g1 was included because the program identified three transcripts belonging to the cluster TRINITY_DN1123_c0. One of them (corresponding to defensin_g2) could be related with gene C0J52_24001 (encoding a hypothetical protein), although we recovered the correct reading frame after the correct placement of the start of the second exon. The other two transcripts showed 100% identity but differed in the alternative splicing of a 453-nt 3′-UTR intron. We considered them isoforms of defensin_g1, a different gene of defensin_g2, because they differed in seven nucleotides (two in the CDS) plus three indels of different sizes in the 3′-UTR. However, such a sequence was not detected in any scaffold sequence.
The highly expressed transcript (TRINITY_DN13842_c0_g1_i1) was apparently derived from the incorrect assembly by TRINITY of the reads of four different loci in the genome with almost identical sequences (defensin_g3 to g6). Three of them were previously annotated with the locus_tag qualifiers C0J52_27569, C0J52_22338 and C0J52_24004. However C0J52_27569 (gene = DEFI_4 in scaffold PYGN01003429) was a tandem of two genes (defensin_g3 and defensin_g4). An assembly gap overlapping with defensin_g3 is probably the reason explaining why a single mRNA expanding both genes was annotated in the genome.
Genes defensin_g7 and defensin_g8 displayed identical CDS sequences but with several differences in the UTR segments of the mRNA sequences. They were placed in scaffolds PYGN01002380 and PYGN01001185, respectively. Only one of them, defensin_g8, was previously annotated as gene C0J52_22336.
Defensin_g9 corresponds to gene C0J52_24005 encoding Phormicin, a 91-amino-acid protein. The transcript analysis revealed that the encoded protein is shorter (71 amino acids) with a signal peptide sequence of 20 amino acids at its amino-terminus (see below). Defensin_g10 was also a Phormicin located in a different scaffold, but only the second exon was present in the genome, with the first exon most likely placed in a contiguous 1-kb assembly gap.
Defensin_g11, g12 and g13 are equivalent to previously annotated genes (Supplementary Tables 2 and 3). Defensin_g14 is present in scaffold PYGN01001185, but most of the sequence of the second exon is absent because of an assembly gap. The CDS sequences of defensin_g15 and C0J52_20459 were identical but transcript analysis of defensin_g15 suggested a two-exon mRNA instead of the three-exon C0J52_20459.
All Defensins showed signal peptides of 18 to 22 amino acids at the N-terminus and the PF01097 (Defensin_2) domain at the C-terminus (see examples of domain organizations at Fig. 1). The length of the amino acid chain ranged from 63 to 81 residues with an average of 72 amino acids. Although some Defensin proteins were identical, the average number of pairwise differences was high (29 amino acids). An inferred maximum likelihood phylogeny showed their distribution in seven clusters (Fig. 2a). A logo of the protein alignment of the Defensin proteins shows the hydrophobic N-terminal sequence as well as the Defensin_2 domain (C-terminus) with the six conserved cysteine residues (Supplementary Fig. 1).
Domain organization in the five types of AMPs of B. germanica. One protein of each class is shown. Orange squares are signal peptides. Red oval corresponds to a glutamine/glutamic acid-rich region. Green ovals are Pfam-A domains PF03769 (Attacin_C). Blue ovals (from top to bottom) are Pfam-A domains PF01097 (Defensin_2), PF11415 (Toxin_37) and PF00304 (Gamma-thionin), respectively.
B. germanica Defensin and Drosomycin protein phylogenies. (a) Maximum likelihood phylogeny of 18 Defensin proteins (derived from transcripts of 16 genes). Model WAG + I with complete deletion. Alignment length 57 sites. Bootstrap replicates 100. Mid-point rooting. (b) Maximum likelihood phylogeny of Drosomycin proteins. Model Dayhoff + G with complete deletion. Alignment length 66 sites. Bootstrap replicates 100. Mid-point rooting. Bootstrap values smaller than 50 are hidden.
A comparison among the levels of transcription of the 16 defensin genes was estimated using a BLASTN strategy based on BLASTN searches with nucleotides 41–190 of each CDS. All 150-nt sequences were different in, at least, one nucleotide, except defensin_g3 and g5 that were identical and the transcription level could not be assigned to a specific gene (Supplementary Table 3). Based on the TPM values estimated by TRINITY and the transcription levels estimated by this BLAST strategy, we observed that in this female adult run, defensin_g15 and g16 (encoding Defensin-like proteins), g9 and g10 (encoding Phormicin) and g1, g2, g3 and g5 (encoding Tenecin-1 proteins) are the most highly expressed defensin genes (Supplementary Table 3).
Using a TBLASTN strategy, defensins transcripts were searched in 45 species covering the order Blattodea26 (Supplementary Table 4). Forty-four species contain defensin transcripts (range 1 to 9).
Termicin AMP genes
Three genes encoding small proteins with the Pfam domain PF11415 are annotated in the genome (Supplementary Table 1). BLASTN searches against the SRR6784710 transcript database gave hits with only two very similar transcripts. The first transcript, TRINITY_DN10017_c0_g1_i1, displayed one single difference with either C0J52_00758 or C0J52_26761 at the CDS sequence but several at the remaining mRNA sequence, suggesting two independent genes in the genome. The second transcript, TRINITY_DN10017_c0_g2_i1, was 100% identical with both CDS and mRNA from C0J52_26762, indicating a third termicin gene. The three encoded proteins are almost identical with a single S/A difference at site 13 (Supplementary Fig. 1). A signal hydrophobic peptide is predicted between amino acids 1 and 19 and the Toxin_37 domain (PF11415) between amino acids 30 and 63 (Fig. 1). Based on the TPM values estimated by TRINITY and the transcription levels estimated by BLASTN (a 150-bp segment covering four polymorphic sites in the termicin CDS), we may conclude that termicin_g3 (C0J52_26762) is the most highly expressed termicin gene (Supplementary Table 5).
Termicin mRNAs were detected in 29 Blattodea species belonging to the different taxonomic families (Supplementary Table 4). Their absence was frequent in species from Corydioidea, suggesting the potential loss of this type of gene, although the lack of expression in these samples may not be ruled out.
Drosomycin AMP genes
Ten genes encoding proteins with the domain Gamma-thionin (PF00304) are annotated in three scaffolds of the B. germanica genome. These antifungal proteins receive the name of Drosomycins. BLASTN searches of the annotated CDS against the SRR6784710 transcript database identified only six transcripts including the complete CDS and two insignificant transcripts covering only a CDS segment.
The comparison of the annotated CDS and those derived from these transcripts revealed that only three annotated genes (C0J52_03170, C0J52_03171 and C0J52_12810) were equivalent to three of these transcripts (the former with 2 nucleotide differences). They were annotated as drosomycin_g2, g3 and g5 (Supplementary Tables 2 and 6). One of the three remaining transcripts, corresponding to drosomycin_g6, could be placed in the genome, with a few nucleotide differences, in a non-annotated segment. Finally, the sequences of the other two transcripts were not detected in the genome, although their CDS sequences were highly similar to C0J52_03170 (with 6 and 8 nucleotide differences). These differences suggest that they are not alleles but independent genes and we annotated them as drosomycin_g1 and g4 (Supplementary Tables 2 and 6).
On the other hand, six annotated genes with locus_tags, C0J52_12811-13 and C0J52_23105-08 were not detected in the adult female transcriptome, but they seem to be expressed in other developmental stages. They were annotated as drosomycin_g7 to g13.
A phylogeny of the 13 Drosomycin proteins showed that defensin_g6 was the most distant gene, while the other 12 genes formed two clusters of six genes each. Genes drosomycin_g1 to g5, expressed in adult females, plus the non-expressed drosomycin_g9 formed one well-supported clade while the other six non-expressed genes formed the other (Fig. 2b).
An estimation of the transcription level revealed that drosomycin_g5 (C0J52_12810) was the gene with the highest expression, with 86.1% of the drosomycin reads for this segment derived from it (Supplementary Table 6).
Twelve out of 13 encoded proteins were 66 amino acids long. Drosomycin_g6 was 71 amino acids long due to the presence in the middle of the protein of additional amino acids derived from two indels (sites 25–26 and 36–38 of the alignment). Among the observed residues, the most remarkable feature in the encoded proteins is the presence of eight conserved cysteines27 (Supplementary Fig. 1). All Drosomycins display a signal hydrophobic peptide at the N-terminus and the PF00304 domain (Gamma-thionin) at the C-terminus (Fig. 1).
Drosomycin mRNAs were detected in 24 Blattodea species but they were absent in species of Isoptera and their close relative Cryptocercus wrighti (Supplementary Table 4). The same fact was detected in the clade Corydioidea, suggesting that termites and other Blattodea may have lost this type of AMP gene.
Attacin AMP genes: attacin-like and blattellicins
Up to four regions with some similarity with the Attacin_C domain (PF03769) were detected in the 47-kb-region spanning the C0J52_26498 gene placed in contig PYGN01001824. After a preliminary analysis of the assembled transcriptome, more than ten mRNA sequences were identified. They resemble complete or partial sequences of mRNAs belonging to two types of attacin genes. The first type includes genes encoding typical Attacin proteins (around 120 amino acids) with a signal peptide at the N-terminus and the Attacin_C domain at the C-terminus, which were named as attacin-like genes. The second type was very different, because it included a long stretch of glutamine/glutamic acid residues. Since they seemed an apparent evolutionary innovation in B. germanica, we called them blattellicins.
Three attacin-like transcripts were detected in the transcriptome (Supplementary Tables 2 and 7). They contained coding sequences of 357–360 nucleotides (118–119 encoded amino acids). They received the names of attacin-like_g1 to attacin-like_g3. The extraction and assembly of the reads for these mRNAs confirmed their existence, but suggested the possibility of a fourth gene. Attacin-like_g3A and attacin-like_g3B display only two differences, the deletion of a 9-nucleotide-segment in the 5′UTR of attacin_g3B and a synonymous difference at CDS position 288 (the inclusion of the sites for the two differences in a read was very infrequent considering that the length of a read is 301 nucleotides). Because there were only two differences and they were unplaced in the genome, we considered that they were alleles of the same gene.
Attacin-like_g1 CDS was relatively similar to attacin-like_g3 CDS with 9–10 differences. However, they were sufficiently different to be considered independent loci. Attacin-like_g2 was the most divergent gene with 85–88 differences and an extra codon against the others. Only the sequences of attacin-like_g1 and g2 were located in the genome (Supplementary Tables 2 and 7).
The annotation of blattellicins was much more complicated. After a preliminary analysis, a long CDS (> 250 codons) with a curious structure was observed. It started with the hydrophobic signal peptide at the N-terminus, followed by a long Glx-rich segment in the middle (> 70 residues, mainly glutamines and glutamic acids) and a C-terminal Attacin domain (Fig. 1).
Up to 13 mRNA transcripts (all of them containing incomplete CDS segments) involving this type of sequences were detected. The main reasons were that the presence of several blattellicin genes and the long Glx-rich regions drastically affected the assembly of the transcriptome. This fact probably took place during the assembly and annotation of B. germanica genome5,6.
The 5′ sequence of a blattellicin CDS was used as query to identify with BLASTN those reads derived from the expression of blattellicin genes in the run SRR6784710. After extraction and assembly, four different starts of blattellicin genes were revealed, with a range of 7 to 18 pairwise nucleotide differences in the 5′ of mRNAs. These four mRNA starts were used to recruit the remaining gene sequences until CDS completion.
Most of the CDS sequence for blattellicin_g1 could be identified in the genome, although around 200-bp were absent due to two assembly gaps (Supplementary Tables 2 and 7). For the others, only the first coding exon of blattellicin_g2 and g4 could be unequivocally assigned to a specific contig segment, although hits for other segments of the CDS were also detected but without 100% identity. No identical sequence to blattellicin_g3 first exon could be identified in the genome. The most feasible explanation is that the four blattellicin genes are present in tandem copies in the genome but their special central repeat structure prevents correct assemblies in either genome or transcriptomes, except if manual inspection of the alignments is performed. Furthermore, variations in Glx codon copy number in the population cannot be ruled out.
We detected that blattellicins were expressed at a higher level than attacin-like genes, with blattellicin_g4 being the most highly expressed in this transcriptome (Supplementary Table 7).
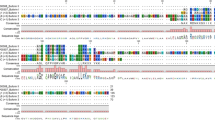
Logos of the protein alignments for the three Attacin-like and the four Blattellicin proteins of B. germanica revealed a small segment of negatively charged amino acids in Attacin-like proteins and a long segment in Blattellicins (Fig. 3).
Attacin mRNAs were detected in most Blattodea species (Supplementary Table 4). Hits for blattellicins did not cover the Glx region but only the attacin_C domain. To understand the evolutionary history of attacin-like and blattellicin genes in B. germanica, we extracted the attacin transcripts from seven Blattellinae TSA projects26 (Symploce sp. AD-2014, Loboptera decipiens, Episymploce sundaica, Ischnoptera deropeltiformis, Paratemnopteryx couloniana, Lobopterella dimidiatipes, Asiablatta kyotensis). These transcriptomes come from adult whole bodies except I. deropeltiformis (without information about developmental stage). They may potentially cover all attacins genes for each genome, although the possibility of genes without expression cannot be discarded. The largest number of attacin genes was three in E. sundaica. Two genes were observed in L. decipiens, Symploce sp. AD-2014 and A. kyotensis, although in the former one of the copies was incomplete and very divergent, probably a pseudogene, while in the latter, the two copies were a few codons incomplete at 5′-end of the CDS. The SRA project was screened for reads covering the start of the CDS and, based on those recovered, one was completed and, in the other, only four codons were missed. The remnant species contained a single gene copy. In addition, in order to use as an outgroup, the only-one-detected in P. americana was extracted.
A phylogeny was performed with a trimmed alignment (103 sites) (Fig. 4). The short length of the sequence alignment prevented high bootstrap values in most nodes and hampered determining with complete confidence the evolutionary history of this gene family. However, several facts are observed from the phylogeny. First, attacin-like genes are the ancestral gene type. Some Blattellinae species contain only one or two genes. In the case of the clade of B. germanica, E. sundaica, L. decipiens and Symploce sp. AD-2014, the duplication of an ancestral attacin-like gene took place before their divergence, resulting in the appearance of attacin-like_g1 and g2 types. Although L. decipiens attacin-like_g1 was not included in the phylogeny, an incomplete and divergent copy of a transcript of this type (GDYK01026461.1) is detected, probably derived from a pseudogenized copy.
Attacin-like and Blattellicin protein phylogeny in Blattellinae. (a) Maximum Likelihood phylogeny of proteins containing Attacin_C domain in the subfamily Blattellinae. Model LG + G with complete deletion. Alignment was trimmed to join the N-terminal signal peptide plus the C-terminal attacin_C domain (length 103 sites). P. americana was used as an outgroup. Bootstrap replicates 100. Bootstrap values smaller than 50 are hidden. All species names are abbreviated (see codes in the right topology), except Symploce sp. Those without abbreviations are proteins from B. germanica. (b) Taxonomic relationships according to26.
The origin of blattellicins seems to be very recent. Although not supported by a significant bootstrap value, potentially an ancestral attacin-like_g2 type gene was duplicated and one of the copies, after a fast evolution, generated blattellicins. The duplication took place prior to the divergence of E. sundaica and B. germanica. The protein in the former is apparently a pre-Blattellicin, including some of the new characteristics of Blattellicins, such as large size (182 residues) and a few extra amino acids at the C-terminus (RK in B. germanica and GKGK in E. sundaica). However, the main characteristic of Blattellicins, the long poly-Glx region, is absent, although E. sundaica pre-Blattellicin includes a seven-glutamic-acid track (with an A in the middle) close to the start of the attacin domain.
AMP expression in B. germanica
To determine the expression of AMP genes in B. germanica tissues, developmental stages or sexes, we selected the CDS of 17 AMP gene types (defensin_g2, g3, g7, g9, g11, g13 and g15; termicin_g1; drosomycin_g1, g5, g6, g11 and g12; attacin-like_g1 and g2; blattellicin_g1 and g4). They are sufficiently different to avoid important cross results among those selected of the same group. However, due to the high similarity of the CDS of some genes from the same family, the obtained values showed the expression of the sets of genes with almost identical sequences (for example, the three termicin genes or attacin-like_g1 and g3).
Expression levels were estimated with a BLASTN strategy as number of hits/Gb of SR experiment (Supplementary Table 8). The heatmap analysis of 28 whole body SR experiments corresponding to samples from different developmental stages (Fig. 5) revealed several conclusions. First, adult females displayed high expression of most AMP genes, although the most relevant was the highest expression of blattellicin_g1 and g4. Some drosomycins were also highly expressed, specially, drosomycin_g5. The expression of some genes was connected with development (see, for example, the absence of expression of drosomycin g11 and g12 in adult females but high expression in nymphs). Among defensins, the most highly expressed during most developmental stages were defensin_g9 and g15. The expression of defensin g2 and g3 was higher in adult females than in nymphs. Termicin_g1 displayed low expression in nymphs and adults. Attacin-like genes were also expressed in adult females, with attacin-like_g1 values higher than those of attacin-like_g2, which was in agreement with previously described results (Supplementary Table 7), also considering that the detected hits for attacin-like_g1 probably comes from g1 and g3 genes.
Gene expression of 17 AMP genes in whole bodies of B. germanica. Heatmap analysis illustrating the abundance of transcripts for 17 selected AMP genes in 28 Sequence Read experiments corresponding to whole bodies from diverse developmental stages of B. germanica with indication, in some cases, of the sample sex. Values were estimated as the quotient between the number of reads producing a hit with an e-value smaller than 1.0E−40 (using the complete CDS sequences as queries in BLASTN searches) and the size in Gb of the SR experiment.
In general, AMP genes display an increase in expression as development progresses to adult forms. Unfortunately, no SR experiment for exclusively adult males was deposited in SRA database, although some mixed males and females samples are reported (Supplementary Table 8).
We also analysed the expression of these 17 AMP genes in some transcriptomic SR experiments in which samples come from one single tissue, a part of the body or a mixture of several tissues (Supplementary Table 8). In general, drosomycin_g5 and defensin_g9 seem to be expressed in most of these samples. In two experiments from male adult heads, several AMP genes were expressed to a relevant level, including defensin_g7 and g9, drosomycin_g5 and attacin-like_g2. In general, the level of expression in these samples is much smaller than those coming from whole bodies. This leads us to propose that other parts of the body different from fat body, ovaries or epidermis are responsible for the high expression levels observed in whole body adult females (Fig. 5).
No expression of blattellicin_g1 and blattellicin_g4 was observed in any tissue or part of the body sample, except an almost undetectable expression in one non-fecundated eggs sample, probably due to contamination with female tissues.
Discussion
Estimating the number of AMP genes in any species is a complex task due to their small sizes, to the evolutionary dynamics of these gene families with frequent gains and losses and with redundancy and neofunctionalization12,14 and to the difficulties of the treatment of repeats in the genome assemblies28. In this work, we have detected in B. germanica an arsenal of genes encoding AMPs larger than initially annotated, including some no yet described (Table 1 and CDS and protein sequences in Supplementary Information). However, we cannot discard the presence of immune taxonomically restricted genes encoding divergent or non-characterized antimicrobial domains, as observed in other species29,30.
When the set of 11 AMP genes of the American cockroach P. americana4 was compared with the AMP gene set annotated in B. germanica, the expansion of some key protein families, such as the anti-fungal drosomycins were described5. A deeper analysis (present study) has revealed the existence of 13 drosomycin and 16 defensin genes. Some are very similar, almost identical, but others display differences in more than 50% of the encoded amino acids (Fig. 2). Two additional families, termicins and attacins, were detected. Three, almost identical, termicin genes are detected in B. germanica in front of two in P. americana. Because of their high identity, two possibilities exist: they are recent duplications, or they evolved by gene conversion. We have divided the gene family of attacins in attacin-like (three genes) and blattellicins (four genes). Although initially two Attacin domains were annotated in Diptera (the Attacin_N and the Attacin_C, at each end of the proteins), Attacin protein alignments of distant insects suggested that Attacin_N is an artefact12 and that Attacin_C is the only domain common to Attacins. Both B. germanica Attacin-like proteins and Blattellicins contain this domain, although these proteins differ drastically in size and in the presence of the long Glx-rich region (Fig. 1). Two additional gene types, one AFP (antifungal peptide) and a Pro-rich peptide, detected in P .americana4, seem not to have a counterpart in B. germanica.
The search of the five types of B. germanica AMP genes in TSA projects from Blattodea (Supplementary Table 4) revealed that four of them (defensins, termicins, drosomycins and attacins) were widely distributed against the different taxonomic suborders, superfamilies or families. However, it was relevant the loss of the antifungal drosomycin genes in the termites and the wood roach Cryptocercus, which would be related with the different types of interactions with fungal symbionts acquired during evolution, as the domestication of Termitomyces fungi in Macrotermitinae31. However, this loss may be compensated by the presence of several termicin genes (Supplementary Table 4), potentially displaying strong antifungal activity32. Expansions of the termicin gene family were observed in two termite species affected by fungal contamination33.
By using BLASTP searches against UniProtKB database limited to Insecta (data not shown), we detected Defensin proteins mainly in Blattodea, Coleoptera, Diptera, Hemiptera and Hymenoptera and a few cases in other taxonomic orders. Termicins were restricted to Blattodea. Drosomycins were detected in Blattodea, Coleoptera and Diptera. Similar sequences to Attacins were detected in Blattodea, Diptera, Hemiptera, Orthoptera and Phasmatodea.
It has been suggested that defensin genes were already present in the last common ancestor of all insects12. Attacins were also considered ancient genes and their origin was placed before de divergence of Palaeoptera and the clade Polyneoptera/Holometabola12. The origin of drosomycins is controversial because they have been found in plants and in Ecdysozoa (Arthropoda, Nematoda and Tardigrada) and it was suggested that the ancestor of ecdysozoans acquired this type of gene from plants through a single horizontal gene transfer event34. Finally, termicin genes are apparently an innovation that predated Blattodea divergence.
The microbial targets for each type of B. germanica AMP are not characterized but, based on a CRISPR gene editing study in Drosophila, we know that some AMPs have broad taxonomic ranges and others very narrow10. While two D. melanogaster Drosomycins inhibit the growth of several fungal species, they are inactive against all the bacterial species tested35. Termicins display strong antifungal activity in termites and very low anti Gram-positive bacteria32. Activity assays of Defensins (PF01097) in species belonging to different insect orders (Diptera, Coleoptera, Hymenoptera, etc.) revealed effects mainly on Gram-positive species and in a lesser extent on Gram-negative and fungal species (see Function section in UniProtKB database files). In Drosophila, it was shown than Attacins are mainly affecting anti Gram-negative bacteria36 and they are induced after immune challenge with a Gram-negative pathogenic Serratia37.
An estimation of the isoelectric points (pI) of complete B. germanica AMP proteins reveals that only Defensins and Drosomycins are cationic. Blattellicins and Termicins are anionic while the pI of Attacin-like proteins is around 7. The protein Defensin 2 from the tick Amblyomma hebraeum with a pI 4.44 showed activity against Escherichia coli and Staphylococcus aureus38. Non-cationic Defensins were also detected in some lepidopteran species such as Spodoptera littoralis39, Bombyx mori40 and Galleria mellonella41. In the latter species, anionic peptide 2 (a mature peptide of 60 residues) contains 13.3% and 8.3% of glutamic acid and glutamines, respectively. A recent study showed that this peptide decreases the survival rate of Candida albicans through its action on the cell wall, and bioinformatics analysis suggest that this action is mediated by the presence of amphipathic alpha-helices with exposed positively charged lysine residues, regardless of its anionic character7. Termicin structure from the termite Pseudacanthotermes spiniger displays a hydrophobic face formed by a large aggregate of hydrophobic residues and a hydrophilic face, including three charged residues42. Because anionic B. germanica AMPs contain positively charged amino acids in relevant proportions (Termicins ca. 6%, Blattellicins ca. 13% and Attacin-like ca. 20%), we cannot discard that they contain protein 3D regions with abundance of positively charged residues, or they are able to produce cationic mature peptides after proteolytic cleavage.
The most relevant fact of Blattellicins, the new type of Attacins, is the long Glx-rich region, whose function is unknown but polyglutamine (polyQ) tracts has been detected in some transcription factors such as TATA-binding proteins43, and several human proteins, such as Ataxins or Huntingtin, are involved in diseases related with glutamine expansions. PolyQ containing proteins tend to produce fibrillar aggregates and to bind lipid membranes in a polyQ-length dependent manner44. Prediction of the secondary structure of Blattellicins renders several contiguous alpha-helices covering most of the segment between the signal peptide and the Attacin_C domain (data not shown). The presence of charged amino acids (Glu) may aid in solubility as previously described for polyQ peptides44. An analysis of the amino acid composition of 398 proteins with an Attacin_C domain in insects (UniProtKB database) did not show high percentages (> 10%) of either glutamine or glutamic acid in most of them. High percentages were detected in only 4 proteins and the only relevant sequence was a 578-residues Attacin C domain-containing protein of unknown function from Anopheles stephensi containing long stretches of glutamine residues and some glutamic acids in the proximity of the attacin_C domain (A0A182Y178). AMP gene expression during development is not homogeneous in B. germanica, with genes expressed during the whole development, genes mainly restricted to adults and genes expressed only during nymphal stages (Fig. 5). Previously, the enriched expression in B. germanica adults of genes involved in immune defence such as those containing the Defensin_2 domain was reported45. The level of expression of dorsal during development of Tenebrio molitor, which is involved in the Toll immune pathway, is also variable with the highest levels in young adults46. In D. melanogaster, the expression analysis of seven drosomycin genes in whole bodies along development, showed some genes expressed from larva to adult and others only expressed in some developmental stages35.
Because the number and types of B. germanica analysing tissue-specific transcriptomic projects is reduced (Supplementary Table 8), we do not know which are the main tissues in both nymphal and adult stages where each AMP gene is expressed. A broader study should be performed to discriminate among some potential tissues where components of the insect humoral immune system are known to be expressed46,47,48,49, such as fat body, haemocytes, gut, salivary glands, trachea, Malpighian tubules, and integument. Fat bodies have been reported to display high expression level for some AMP types such as defensins49,50 and attacin51. Defensin genes from adult A. stephensi are expressed in fat body, midgut, haemolymph and salivary glands with the high level of expression at fat body49. On the contrary, a defensin gene from Bemisia tabaci was more highly expressed in adult midgut or salivary glands than in fat bodies52 and a type of attacin gene was highly expressed in adult Malpighian tubules of the stick insect Carausius morosus53. It was also reported constitutive expression of a termicin gene in adult haemolymph and salivary glands from the termite P. spiniger32.
Of special interest is the expression in hindgut, along the development, because the rich and variable microbiota that is acquired after birth may be, somehow, controlled by the host. Moreover, for a correct relationship with Blattabacterium, its ancient endosymbiont, B. germanica could be using one of its AMP genes, expressing it differentially in the fat body bacteriocytes and in the rest of the body, as it is the case in Sitophilus zeamays with coleoptericin A14,54. Because Coleoptericin and Attacin contain a related domain, we can imagine that one of the Attacin-like proteins or the Blattellicins may be in charge of this function. In the louse fly Melophagus ovinus, attacin is one of the genes upregulated in midgut but downregulated in bacteriocytes55, probably related to the relationship with its obligate endosymbiont Arsenophonus melophagi.
Finally, knowing the tissues where Attacin-like proteins and Blattellicins are expressed will require additional RNA-Seq studies including both developmental stages and specific tissues. The expression of two attacin-like genes in adult male heads may be related with the initial parts of the gut, while the absence of Blattellicin synthesis in fat body, ovaries and epidermis (Supplementary Table 8) suggests their synthesis in other tissues or cells, such as the different parts of the gut, Malpighian tubules, salivary glands or haemocytes.
Methods
Sequence data and strategy for AMP gene set revision
The genome sequence of B. germanica (Orlando Normal from American Cyanamid) was obtained from GenBank assembly accession GCA_003018175.1 (WGS Project PYGN01)6. The sequence data of run SRR6784710 (SRX3744248), belonging to B. germanica whole body adult female 5-days old sample45, was used to produce an assembled transcriptome. To integrate the genome annotation and the assembled Trinity transcripts, each transcript was aligned with the genome using BLASTN against B. germanica genome project (GCA_003018175.1). Transcripts were also aligned to scaffold sequences with the program Spidey in the platform UGENE56, to check for the correct splicing, to compare potential alternative spliced transcripts, and to identify partial genes.
Information from available B. germanica transcriptomic projects (Supplementary Table 8) was extracted from NCBI.
A TBLASTN strategy was used to identify transcripts producing hits using B. germanica AMPs as queries (e-value = 0.001) in 44 Blattodea SRA projects plus the annotated mRNAs of Z. nevadensis26 (Supplementary Table 4). Sequences of attacin CDS were obtained from the transcriptome shotgun assemblies of seven Blattellinae TSA projects (Symploce sp. AD-2014, L. decipiens, E. sundaica, I. deropeltiformis, P. couloniana, L. dimidiatipes, A. kyotensis)26.
Transcriptome assembly
The transcriptome of B. germanica was assembled from the SRA project SRR6784710. Fastq files obtained using SRA toolkit fastq-dump tool (http://ncbi.github.io/sra-tools/) were filtered with PRINSEQ57 to obtain sequences with a mean quality of 20 Phred and a minimum length of 100 bases. Illumina adapters were removed using cutadapt58. The assembly was performed with de novo Trinity25 using default parameters.
Transcriptome level estimations
Although, TFM (transcripts per million transcripts) values were estimated for the transcriptome assembled with de novo Trinity (Supplementary Tables 3, 5, 6 and 7), because the integrated revision of genome annotation and transcriptome rendered a slightly different set of AMP genes, we developed an additional strategy. To estimate the level of expression of genes in adult females, BLASTN searches were performed using CDS segments of 150 nucleotides length as queries against SRX3744248, and the number of hits covering 100% of the query with 100% identity were filtered and counted. In drosomycins and attacins/blattellicins, we use the first 150 nucleotides of each CDS but, in the defensins and termicins, we selected a different segment trying to discriminate the maximum number of genes but, in a few cases, it was not possible (Supplementary Tables 3, 5, 6 and 7). For each gene, the expression value was calculated relative to its type of AMP gene family, as a percentage over the total number of hits for the set of genes of its type (Supplementary Tables 3, 5, 6 and 7).
To estimate the level of expression in SR experiments of whole bodies during development or in tissues or body parts (Fig. 5 and Supplementary Table 8), BLASTN searches were performed using the complete CDS of 17 AMP genes as queries against each SR experiment. Gene expression was estimated as the quotient between the number of reads producing a hit with an e-value smaller than 1.0E−40 (maximum of 5000 results) and the size in Gb of each SR experiment. An additional normalization (hits/Gb and length CDS) was calculated, but, although the long blattellicins displayed comparatively lower levels, they were already the highly expressed AMP genes in whole body adult females. Because several AMP genes belonging to the same family are very similar, the reported expression values may include hits belonging to additional genes (for example, the expression value of termicin_g1 is probably covering the expression of the three termicin genes). In general, the 17 AMP genes were selected based on its position in the AMP phylogenies, trying to select one gene for each phylogenetic group. In general, the selected genes, in each group, were those with the highest expressions in adult females, except drosomycin_g11 and g12 without expression (Supplementary Tables 3, 5, 6 and 7). Heatmaps were plotted using R 3.6.059 and package lattice60 (v0.20-41).
Sequence alignment and phylogenetics
BLAST searches were performed at the NCBI server or locally with BLAST+ software61. Multiple sequence alignments were performed in MEGA762 and in Unipro UGENE56 with the program MUSCLE63. For phylogenetic analyses, AMP protein alignments were manually revised according to the domain alignments generated with HMMER (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan). The evolutionary history was inferred by using the Maximum Likelihood method based on the estimated best protein evolutionary model. For Attacins (including Blattellicins), the phylogeny was inferred using a trimmed alignment corresponding to the concatenation of the first 22 amino acids (20 amino acids of the signal peptide plus two extra amino acids, being the last one proline) plus 81 sites of the C-terminal attacin domain (domain sequence lengths range 74–81).
Domain organization and signal peptide
The modular structure of AMP proteins (PFAM domains and signal peptides) were identified online (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan). Domain architecture images were generated in PFAM (https://pfam.xfam.org/generate_graphic). Logos of aligned AMP proteins were generated with WebLogo 3 (http://weblogo.threeplusone.com).
Data availability
B. germanica CDS and protein sequences of the described AMPs may be found in Supplementary Information.
References
Steiner, H. et al. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292, 246–248 (1981).
UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Kang, X. et al. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 6, 148 (2019).
Li, S. et al. The genomic and functional landscapes of developmental plasticity in the American cockroach. Nat. Commun. 9, 1008. https://doi.org/10.1038/s41467-018-03281-1 (2018).
Harrison, M. C. et al. Expansions of key protein families in the German cockroach highlight the molecular basis of its remarkable success as a global indoor pest. J. Exp. Zool. B. Mol. Dev. Evol. 330, 254–264. https://doi.org/10.1002/jez.b.22824 (2018).
Harrison, M. C. et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2, 557–566. https://doi.org/10.1038/s41559-017-0459-1 (2018).
Sowa-Jasiłek, A. et al. Antifungal activity of anionic defense peptides: Insight into the action of Galleria mellonella anionic peptide 2. Int. J. Mol. Sci. 21, 1912. https://doi.org/10.3390/ijms21061912 (2020).
Koehbach, J. Structure-activity relationships of insect defensins. Front. Chem. 5, 45. https://doi.org/10.3389/fchem.2017.00045 (2017).
Hillyer, J. F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 581, 2–118. https://doi.org/10.1016/j.dci.2015.12.006 (2016).
Hanson, M. A. et al. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife 8, e44341. https://doi.org/10.7554/eLife.44341 (2019).
Lazzaro, B. P., Zasloff, M. & Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 368, eeau5480. https://doi.org/10.1126/science.aau5480 (2018).
Mylonakis, E., Podsiadlowski, L., Muhammed, M. & Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Phil. Trans. R. Soc. B. 371, 20150290. https://doi.org/10.1098/rstb.2015.0290 (2016).
Oakeson, N. F. et al. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol. Evol. 6, 76–93. https://doi.org/10.1093/gbe/evt210 (2014).
Login, F. H. et al. Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365 (2011).
Mergaert, P., Kikuchi, Y., Shigenobu, S. & Nowack, E. C. M. Role of antimicrobial peptides in controlling symbiotic bacterial populations. Trends Microbiol. 25, 703–712 (2017).
López-Sánchez, M. J. et al. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 5, e1000721. https://doi.org/10.1371/journal.pgen.1000721 (2009).
Neef, A. et al. Genome economization in the endosymbiont of the wood roach Cryptocercus punctulatus due to drastic loss of amino acid synthesis capabilities. Genome Biol. Evol. 3, 1437–1448. https://doi.org/10.1093/gbe/evr118 (2011).
Sabree, Z. L., Kambhampati, S. & Moran, N. A. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. 106, 19521–6. https://doi.org/10.1073/pnas.0907504106 (2009).
Sabree, Z. L. et al. Genome shrinkage and loss of nutrient-providing potential in the obligate symbiont of the primitive termite Mastotermes darwiniensis. App. Environ. Microbiol. 78, 204–210 (2012).
Patiño-Navarrete, R., Moya, A., Latorre, A. & Peretó, J. Comparative genomics of Blattabacterium cuenoti: The frozen legacy of an ancient endosymbiont genome. Genome Biol. Evol. 5, 351–61. https://doi.org/10.1093/gbe/evt011 (2013).
Patiño-Navarrete, R. et al. The cockroach Blattella germanica obtains nitrogen from uric acid through a metabolic pathway shared with its bacterial endosymbiont. Biol. Lett. 10, 20140407 (2014).
Carrasco, P. et al. Succession of the gut microbiota in the cockroach Blattella germanica. Int. Microbiol. 17, 99–109. https://doi.org/10.2436/20.1501.01.212 (2014).
Pérez-Cobas, A. E. et al. Diet shapes the gut microbiota of the omnivorous cockroach Blattella germanica. FEMS Microbiol. Ecol. 91, 1–14. https://doi.org/10.1093/femsec/fiv022 (2015).
Rosas, T. et al. Rifampicin treatment of Blattella germanica evidences a fecal transmission route of their gut microbiota. FEMS Microbiol Ecol. 94, 1–47 (2018).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–52. https://doi.org/10.1038/nbt.1883 (2011).
Evangelista, D. A. et al. An integrative phylogenomic approach illuminates the evolutionary history of cockroaches and termites (Blattodea). Proc. R. Soc. Lond. B B286, 20182076. https://doi.org/10.1098/rspb.2018.2076 (2019).
Gao, B. & Zhu, S. The drosomycin multigene family: Three-disulfide variants from Drosophila takahashii possess antibacterial activity. Sci. Rep. 6, 1–12 (2016).
Tørresen, , et al. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 47, 10994–11006 (2019).
Khalturin, K., Hemmrich, G., Fraune, S., Augustin, R. & Bosch, T. C. G. More than just orphans: Are taxonomically-restricted genes important in evolution?. Trends Genet. 25, 404–413 (2009).
Sackton, T. B. Comparative genomics and transcriptomics of host–pathogen interactions in insects: Evolutionary insights and future directions. Curr. Opin. Insect. Sci. 31, 106–113 (2019).
Poulsen, M. et al. Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc. Natl. Acad. Sci. 111, 14500–14505. https://doi.org/10.1073/pnas.1319718111 (2014).
Lamberty, M. et al. Insect immunity. Constitutive expression of a cysteine-rich antifungal and a linear antibacterial peptide in a termite insect. J. Biol. Chem. 276, 4085–4092. https://doi.org/10.1074/jbc.M002998200 (2001).
Xu, P., Shi, M., Lai, R. & Chen, X. X. Differences in numbers of termicins expressed in two termite species affected by fungal contamination of their environments. Genet. Mol. Res. 11, 2247–2257. https://doi.org/10.4238/2012.May.10.2 (2012).
Zhu, S. & Gao, B. Nematode-derived drosomycin-type antifungal peptides provide evidence for plant-to-ecdysozoan horizontal transfer of a disease resistance gene. Nat. Commun. 5, 3154. https://doi.org/10.1038/ncomms4154 (2014).
Tian, C. et al. Gene expression, antiparasitic activity, and functional evolution of the drosomycin family. Mol. Immunol. 45, 3909–3916. https://doi.org/10.1016/j.molimm.2008.06.025 (2008).
Wang, L. N., Yu, B., Han, G. Q. & Chen, D. W. Molecular cloning, expression in Escherichia coliof Attacin A gene from Drosophila and detection of biological activity. Mol. Biol. Rep. 37, 2463–2469. https://doi.org/10.1007/s11033-009-9758-1 (2010).
Hanson, M. A., Hamilton, P. T. & Perlman, S. J. Immune genes and divergent antimicrobial peptides in flies of the subgenus Drosophila. BMC Evol. Biol. 16, 228. https://doi.org/10.1186/s12862-016-0805-y (2016).
Lai, R., Lomas, L. O., Jonczy, J., Turner, P. C. & Rees, H. H. Two novel non-cationic defensin-like antimicrobial peptides from haemolymph of the female tick Amblyomma hebraeum. Biochem. J. 379, 681–685. https://doi.org/10.1042/bj20031429 (2004).
Seufi, A. M., Hafez, E. E. & Galal, F. H. Identification, phylogenetic analysis and expression profile of an anionic insect defensin gene, with antibacterial activity, from bacterial-challenged cotton leafworm Spodoptera littoralis. BMC Mol. Biol. 12, 47. https://doi.org/10.1186/1471-2199-12-47 (2011).
Wen, H. et al. Sequence structure and expression pattern of a novel anionic defensin-like gene from silkworm (Bombyx mori). Mol. Biol. Rep. 36, 711–716. https://doi.org/10.1007/s11033-008-9233-4 (2009).
Cytryńska, M., Mak, P., Zdybicka-Barabas, A., Suder, P. & Jakubowicz, T. Purification and characterization of eight peptides from Galleria mellonella immune hemolymph. Peptides 28, 533–546. https://doi.org/10.1016/j.peptides.2006.11.010 (2007).
Da Silva, P. et al. Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger. Protein Sci. 12, 438–446 (2003).
Purrello, M. et al. Physical mapping at 6q27 of the locus for the TATA box-binding protein, the DNA-binding subunit of TFIID and a component of SL1 and TFIIIB, strongly suggests that it is single copy in the human genome. Genomics 22, 94–100 (1994).
Adegbuyiro, A., Sedighi, F., Pilkington, A. W., Groover, S. & Legleiter, J. Proteins containing expanded polyglutamine tracts and neurodegenerative disease. Biochemistry 56, 1199–1217 (2017).
Ylla, G., Piulachs, M. D. & Bellés, X. Comparative transcriptomics in two extreme neopterans reveals general trends in the evolution of modern insects. iScience 4, 164–179. https://doi.org/10.1016/j.isci.2018.05.017 (2018).
Keshavarz, M. et al. TmDorX2 positively regulates antimicrobial peptides in Tenebrio molitor gut, fat body, and hemocytes in response to bacterial and fungal infection. Sci. Rep. 9, 16878 (2019).
Keshavarz, M., Jo, Y. H., Edosa, T. T. & Han, Y. S. Tenebrio molitor PGRP-LE plays a critical role in gut antimicrobial peptide production in response to Escherichia coli. Front. Physiol. 11, 666 (2020).
Senger, K., Harris, K. & Levine, M. GATA factors participate in tissue-specific immune responses in Drosophila larvae. Proc. Natl. Acad. Sci. 103, 15957–15962 (2006).
Das De, T. et al. Interorgan molecular communication strategies of “local” and “systemic” innate immune responses in mosquito Anopheles stephensi. Front. Immunol. 9, 1–17. https://doi.org/10.3389/fimmu.2018.00148 (2018).
Liu, S.-H. et al. Antimicrobial peptide gene cecropin-2 and defensin respond to peptidoglycan infection in the female adult of oriental fruit fly, Bactrocera dorsalis (Hendel). Comp Biochem. Physiol. Part B Biochem. Mol. Biol. 206, 1–7. https://doi.org/10.1016/j.cbpb.2017.01.004 (2017).
Shelomi, M., Lin, S. S. & Liu, L. Y. Transcriptome and microbiome of coconut rhinoceros beetle (Oryctes rhinoceros) larvae. BMC Genomics 20, 957. https://doi.org/10.1186/s12864-019-6352-3 (2019).
Wang, Z.-Z., Shi, M., Ye, X. Q., Chen, M.-Y. & Chen, X. X. Identification, characterization and expression of a defensin-like antifungal peptide from the whitefly Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Insect Mol. Biol. 22, 297–305. https://doi.org/10.1111/imb.12021 (2013).
Shelomi, M. . De. novo transcriptome analysis of the excretory tubules of Carausius morosus (Phasmatodea) and possible functions of the midgut ‘appendices’. PLoS ONE 12, e0174984. https://doi.org/10.1371/journal.pone.0174984 (2017).
Masson, F., Zaidman-Rémy, A. & Heddi, A. Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Phil. Trans. R. Soc. B 371, 20150298 (2016).
Husnik, F., Hypsa, V. & Darby, A. Insect—symbiont gene expression in the midgut bacteriocytes of a blood-sucking parasite. Genome Biol. Evol. 12, 429–442. https://doi.org/10.1093/gbe/evaa032 (2020).
Okonechnikov, K., Golosova, O. & Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. https://doi.org/10.1093/bioinformatics/bts091 (2012).
Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10 (2011).
R Core Team. R: A language and environment for statistical computing. (2019).
Sarkar, D. Lattice: Multivariate Data Visualization with R (Springer, New York, 2008).
Camacho, C. et al. BLAST+: Architecture and Applications. BMC Bioinform. 10, 1–9 (2009).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Edgar, R. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Acknowledgements
This work was funded by European Regional Development Fund (ERDF) and Ministerio de Ciencia, Innovación y Universidades (Spain) (PGC2018-099344-B-I00) and Generalitat Valenciana (Prometeo/2018/A/133). MMB is granted with a FPU fellowship from the Ministerio de Ciencia, Innovación y Universidades (Spain) (FPU15-01203).
Author information
Authors and Affiliations
Contributions
F.J.S. and A.L. designed the aims of the work. F.J.S. designed the experiments. F.J.S. and M.M.-B. performed the experiments. F.J.S. wrote the manuscript with inputs from A.L., C.G-F. and M.M.-B. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Silva, F.J., Muñoz-Benavent, M., García-Ferris, C. et al. Blattella germanica displays a large arsenal of antimicrobial peptide genes. Sci Rep 10, 21058 (2020). https://doi.org/10.1038/s41598-020-77982-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77982-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.