Abstract
Systemic lupus erythematosus (SLE) flares elicit progressive organ damage, leading to disability and early mortality. This study evaluated clinical and immunologic factors associated with impending flare in the Biomarkers of Lupus Disease study. Autoantibodies and 32 soluble mediators were measured by multiplex assays, immune pathway activation by gene expression module scores, and immune cell subset frequencies and activation states by flow cytometry. After providing baseline samples, participants received transient steroids to suppress disease and were followed until flare. Flare occurred early (within 60 days of baseline) in 21 participants and late (90–165 days) in 13. At baseline, compared to the late flare group, the early flare group had differential gene expression in monocyte, T cell, interferon, and inflammation modules, as well as significantly higher frequencies of activated (aCD11b+) neutrophils and monocytes, and activated (CD86hi) naïve B cells. Random forest models showed three subgroups of early flare patients, distinguished by greater baseline frequencies of aCD11b+ monocytes, or CD86hi naïve B cells, or both. Increases in these cell populations were the most accurate biomarkers for early flare in this study. These results suggest that SLE flares may arise from an overlapping spectrum of lymphoid and myeloid mechanisms in different patients.
Similar content being viewed by others
Introduction
Systemic lupus erythematosus (SLE) is a chronic, debilitating autoimmune disease that causes irreversible organ damage, contributing to diminished quality of life and early mortality1,2. Most SLE patients experience periods of relatively quiescent disease punctuated with periods of increased clinical activity3. Because disease flares and the major immunosuppressants used to subdue disease activity can both cause irreparable damage1, the frequency and severity of flares are important prognostic indicators for long term SLE outcomes4,5,6. In patients receiving standard-of-care treatments, rates of flare range from 0.24 to 1.8 flares per person-year5,6,7. In the phase III belimumab trials, 19.4% of patients who took steroids at baseline in addition to other standard treatments developed a flare over a 1-year period7.
Current clinical and laboratory instruments for forecasting disease flares have some utility but remain inadequate. Low complement C3 levels and rising anti-dsDNA have been useful as predictors of flare in the subset of patients with serologically active disease8,9,10,11,12,13,14,15. Cell-bound complement activation products have been correlated with SLE disease activity in longitudinal studies of patients with active disease and elevated complement activity at baseline16, but not all SLE patients have elevated cell-bound complement activation products17. Another proposed flare predictor, elevated frequency of CD27hi plasma cells, was later shown to have a stronger association with infection than with disease flares9.
Currently available markers do not always predict impending flare18,19,20,21,22. If the timing of flares can be anticipated, it may be possible to optimize treatment to prevent flares while also sparing patients from unnecessary toxic treatments. Thus, predicting disease flares could reduce morbidity and improve early mortality rates in SLE patients. Being able to identify SLE patients with an increased risk of imminent flare would also help in the design of discriminatory clinical trials. Molecular mechanisms of disease flare are also largely unknown, and identification of such mechanisms may be important for therapeutic selection in SLE, as well as development of novel targeted therapies. Therefore, the goal of this study was to identify potential mechanisms of rapid flare after disease suppression through characterization of baseline soluble mediators, immune cell phenotypes, and gene expression patterns.
Results
Characteristics of patients with early or late flare
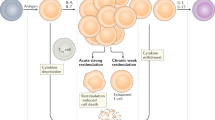
All evaluated patients had active, but not organ threatening, SLE at baseline and clinically significant improvement after receiving one to four bolus depomedrol injections (160 mg each; ≤640 mg total). Background immunosuppressants were withdrawn and patients serially followed until worsening of disease activity associated with clinician’s intention to treat (flare visit) as described23. Of 41 participants, 40 (97.6%) had a flare within 6 months. Of these, 21 had a flare within 60 days (early flare; median time-to-flare of 38 days), and 13 had a flare ≥90 days from baseline (late flare; median time-to-flare of 109 days) (Fig. 1, Table 1). Early flare was more common in African-American participants than others. Other demographic characteristics were similar between the early and late flare groups, as were the ACR criteria; baseline immunosuppressant usage (Table 1); baseline, maximally suppressed and flare disease activity (Supplementary Fig. S1); clinical lab values (Supplementary Table S1); and prevalence and levels of antinuclear autoantibodies measured at baseline (Supplementary Table S2). The early and late flare groups showed no significant differences in the number of steroid injections required to achieve clinical improvement (early flare: median 2, range 2–6; late flare: median 3, range 2–4; p = 0.126).
Identification of patients with early or late flare after steroid-induced disease suppression. (a) Samples and data from this study were obtained from the previously published Biomarkers of Lupus Disease (BOLD) clinical study23. (b) Patients were assigned to three groups based on time to flare during the BOLD study: early flare (<60 days; red; n = 21), intermediate flare (between 60 and 90 days; orange; n = 6), and late flare (90–165 days; blue; n = 13). The current study compared the early flare group vs. the late flare group. IS: immunosuppressant therapy; SOC: standard of care.
Distinct molecular signatures in early and late flare groups at baseline
To identify molecular patterns that preceded early flares, we analyzed baseline samples for transcriptional modules that were previously associated with immune modulation24,25. The early flare group exhibited an elevated interferon signature (M3.4, M5.12), a slightly elevated inflammation signature (M4.6, M5.7; Fig. 2), and a suppressed T cell signature (M4.1, M4.15). The late flare group exhibited a suppressed inflammation signature (M3.2, M4.2, M4.6, M4.13, M5.1, M5.7), a slightly decreased monocyte signature (M4.14), and a slightly elevated T cell signature (Fig. 2). These differences in molecular pathway signatures suggest that the timing of SLE flares may be influenced by functional characteristics in the myeloid and lymphoid compartments.
Transcriptional modules at baseline in SLE patients with early or late flare. (a,b) Activation of transcriptional modules was determined at baseline and compared between SLE patients with early flare (a; n = 21) or late flare (b; n = 13) versus healthy controls. Each box marked with a colored square represents a module, and the color of the square indicates the primary function of the module, as shown at the bottom of the figure. The size of each circle represents the absolute value of the module score. The color represents an increase (red circles; positive scores) or decrease (blue circles; negative scores) in the pathway, in patients compared to controls, as shown at right. P values were determined by non-parametric test. (c) The radar plot summarizes differences between the module scores in the early vs. late flare groups.
Baseline plasma levels of IFN-γ, TNFRII and TNFRI in patients with early or late flare
To further explore immune pathways that may influence time to flare, we compared 32 cytokines, chemokines, adhesion molecules, and soluble tumor necrosis factor receptors (TNFR) at baseline. The early flare group had lower levels of IFN-γ compared to the late flare group, along with slightly lower concentrations of TNFRI and TNFRII (Supplementary Table S3).
Frequencies of activated naïve B cells (CD86hi), neutrophils (activated CD11b+), and monocytes (activated CD11b+) preceding early or late flare
Alterations in cellular profiles have been associated with SLE disease activity and immune dysregulation. Therefore, cellular profiles were assessed by flow cytometry (Supplementary Tables S4, S5). Expression of activated CD11b (aCD11b), determined by a conformation-specific antibody, was used as an indicator of neutrophil and monocyte activation. Patients with an early flare had significantly higher frequencies of activated neutrophils and monocytes compared to patients with a late flare (Fig. 3, Supplementary Table S5). In addition to the altered myeloid compartment, the early flare group had a significantly elevated frequency of activated naïve B cells (CD86hi) compared to the late flare group (Fig. 3, Supplementary Table S5). These results suggest that early flares were preceded by dysregulation in activation of the lymphoid and/or myeloid compartments.
Frequencies of aCD11b+ monocytes and CD86hi naïve B cells distinguish SLE patients with early or late flare in random forest modeling. (a) Baseline frequencies of activated CD11b positive (aCD11b+) monocytes (top) and CD86high B cells (bottom) were quantified by flow cytometry in patients with early flare (n = 21) or late flare (n = 13) after steroid-induced disease suppression. For both comparisons, p < 0.05 by Mann-Whitney U test. (b) Random forest modeling with cellular, clinical, cytokine, and transcriptional panels identified late flare patients and three subgroups of early flare patients. The random forest model proximity matrix is shown as a multi-dimensionally reduced plot, where each point represents a patient, and the distance between two points represents dissimilarity between patients. (c) The variables included in the final random forest model for each independent panel (cellular, clinical, cytokine, and genetic module) are shown in a radial plot. Variables are grouped according to their involvement in the lymphoid, complement/cell adhesion, or myeloid system. Lines represent normalized values for the late flare group and the three early flare subgroups, as indicated.
Three subsets of early flare patients differentiated by elevated frequencies of activated monocytes, activated naïve B cells, or both
Because of the complexity of immune dysregulation in SLE, characterizing pathways related to imminent clinical flares requires a multifactorial model capable of handling various data types (e.g. categorical and continuous) and detecting interactions between variables. The random forest classification algorithm distills large, multifactorial datasets down to the handful of variables that are most informative as independent classifiers. Therefore, to better understand the immune pathways that preceded early flares, we performed random forest modeling with each panel of variables (clinical, cellular phenotypes, expression modules, and soluble mediators) to develop four independent models for early vs. late flare (Table 2).
Of these baseline models for early flare, the model based on cellular phenotypes (n = 30) was the most reliable (86.4 ± 1.4% accuracy). In this model, higher frequencies of activated (aCD11b+) monocytes (>0.2%), or activated (CD86hi) naïve B cells (>2.3%), or both were the strongest markers of early flare (Fig. 3, Table 2). This model revealed three subsets of patients within the early flare group: (1) patients with high frequencies of aCD11b+ monocytes and normal frequencies of CD86hi naïve B cells, (2) patients with high frequencies of CD86hi naïve B cells and normal frequencies of aCD11b+ monocytes, and (3) patients with increased frequencies of both cell populations (Fig. 3). Although molecularly heterogeneous, clinical features were similar across the three subsets of patients with early flare (Supplementary Table S6). Within the subgroups of early flare patients, increases in aCD11b+ monocytes and CD86hi naïve B cells corresponded to higher scores in the M4.10 B cell and M4.14 monocyte correlated gene expression modules, respectively (Figs 3, S2).
The clinical panel produced the second most accurate independent model (79.31 ± 0.01% accuracy), based on complement C3 levels (Table 2). The model based on soluble mediators reached 72.95 ± 0.60% accuracy, based on levels of the CD11b receptor ICAM-1 and the IFN-γ-associated mediator IP-10 (Table 2, Supplementary Fig. S2). Levels of ICAM-1 inversely correlated with the frequency of activated aCD11b+ monocytes (Supplementary Fig. S2). The model based on co-expression module scores reached 66.67% ± 0.01% accuracy, based on the M5.1 inflammation, M4.14 monocyte, and M4.1 T cell module scores.
Finally, in a comprehensive prediction model using only the top biomarkers from each panel, aCD11b+ monocytes and CD86hi naïve B cells frequencies were the strongest predictors of early flare. The comprehensive model outperformed the models based on gene expression modules or soluble mediators alone (88.67 ± 2.69% accuracy; Table 2). However, the comprehensive model was limited by missing values, and underperformed against the cellular phenotypes model. Therefore, elevated frequencies of aCD11b+ monocytes, CD86hi naïve B cells, or both were the most reliable and accurate biomarkers for early flare in this study. These results indicate that SLE flares may follow dysregulation of lymphoid, myeloid, or both pathways.
Discussion
Although enhanced diagnostic tools and increased use of immunosuppressive therapies have reduced SLE-related mortality in the past fifty years, organ damage caused by disease flares and aggressive treatment continues to impede quality of life and long-term survival1,26,27,28. The current study has identified baseline molecular and cellular signatures that distinguish between patients with early and late flare after steroid-induced disease suppression, and reveal multiple pathways preceding flare in these patients.
As expected in a heterogenous disease such as SLE, no single biomarker distinguished all patients with early flare. In the most accurate model, frequencies of activated (aCD11b+) monocytes, or CD86hi naïve (CD19+/CD27−) B cells, or both were sufficient to distinguish patients who had an early flare from those with delayed flare. These variables also defined three distinct subsets of patients with early flare, suggesting that activity of myeloid cells and lymphocytes could contribute, either separately or synergistically, to imminent flares in different subsets of SLE patients. Therefore, an overlapping spectrum of immune dysregulation may lead to SLE flares in different patients. The identification of biomarkers associated with early flare in this study has potential implications for disease monitoring and treatment optimization. This warrants confirmation in a larger population, with detailed immunological analyses of samples collected before, during, and after steroid treatment.
In one patient subset, early flares were preceded by high frequencies of activated (aCD11b+) monocytes, elevated monocyte module scores, and decreased plasma concentrations of ICAM-1, a CD11b receptor. This is consistent with other data supporting an important role for myeloid cells in SLE pathophysiology29,30 and a specific association of CD11b with disease activity31. Functional variants in ITGAM, the gene encoding CD11b, strongly associate with SLE in case-control studies32 and with lupus nephritis, discoid rash, and immunologic manifestations in case-only analyses33. In murine models, activated renal macrophages are a prominent feature of lupus nephritis, corroborating an association between macrophage activation and disease severity34. In addition, CD11b is a component of complement receptor 3, which recognizes iC3b-coated immune complexes and initiates stimulatory pathways in monocytes and neutrophils35,36,37,38.
In another subset of early flare patients, elevated frequencies of activated CD86hi/CD19+/CD27− naïve B cells and increased B cell module scores suggest that flares in some individuals are preceded by perturbed B cell homeostasis. Indeed, abnormal expansion of activated B cells in SLE patients has been correlated with disease activity39,40. Finally, the third subset of early flare patients exhibited increases in both aCD11b+ monocytes and CD86hi B cells at baseline, suggesting that these pathways may act in concert to elicit flares in some patients.
The interferon modules (M1.2, M3.4, and M5.12) were elevated in all groups of SLE patients in this study compared to healthy controls. However, patients with early flare showed a further increase compared to patients with late flare, suggesting a continuum of interferon dysregulation. In addition, different interferon types induce different immune responses and may affect the efficacy of glucocorticoids25,41,42,43. In this study, patients with late flare had slightly higher levels of IFN-γ than patients with early flare. IFN-γ is associated with regulatory pathways in addition to its role in monocyte and B cell activation, and it is possible that production of IFN-γ by regulatory cells would suppress flares in some patients. Alternatively, a slight increase in IFN-γ may arise from occult infection, Th1 cells, or other sources. Therefore, additional studies are needed to determine the source of IFN-γ and to confirm its influence on lupus flares.
In conclusion, this study identified immune variables that distinguish SLE patients with rapid flare after transient corticosteroid-induced disease suppression, suggesting that flares may arise through immune pathways involving activation of naïve B cells, monocytes or both. The ability to identify patients at risk of impending flare could optimize the timing of disease suppression therapy and contribute to more effective and efficient clinical trial designs.
Methods
Patients and samples
This study was approved by the Oklahoma Medical Research Foundation Institutional Review Board and conducted in accordance with the Helsinki Declaration. Written informed consent was obtained prior to study-specific procedures.
These experiments used data and samples obtained at the baseline visit of the Biomarkers of Lupus Disease (BOLD) study (NCT00987831), which recruited SLE patients with moderate to severe, but non-organ threatening disease23. Upon enrollment (baseline), background immunosuppressants were withdrawn, with the option to continue hydroxychloroquine and up to 10 mg/day prednisone or equivalent steroid. Patients received transient steroid injection(s) until disease activity was reduced (improving visit), and were closely followed until the first signs of a flare, at which time they were immediately treated (Fig. 1, Supplementary Methods).
Clinical evaluations included the SLE disease activity index (SLEDAI), British Isles Lupus Assessment Group (BILAG) 2004 Index, physician global assessments (PGA)44,45,46,47, complete blood counts (CBC) with differential, blood chemistries, urine analysis, and clinical-serologic markers of SLE (Supplementary Table S1). Baseline peripheral blood specimens, peripheral blood mononuclear cells (PBMCs), and PAXgene tubes were collected as described (Supplementary Methods and23).
Modular analysis of gene expression molecular signatures
mRNA was isolated from whole blood baseline samples of 21 SLE patients and 15 healthy controls from the BOLD study. mRNA expression in globin-depleted peripheral blood was measured with the HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, California). Quality control was performed with Illumina GenomeStudio Version 2011.1 (Illumina, San Diego, California) per manufacturer protocol.
Gene expression data were analyzed using R version 3.3.2. Background-subtracted expression data were log2 transformed and normalized with the rankinvariant method using the lumiR package48. System-based modular analysis at the group level and the individual level was performed using second generation modular frameworks as described24,25. Individual-level based modular scores were used for subsequent Random Forest prediction models.
Soluble mediator and autoantibody assessments
Antinuclear antibodies (dsDNA, chromatin, ribosomal P, 52 and 60 kD Ro/SS-A, La/SS-B, Sm, Sm/RNP, RNP A and 68 kD RNP, Scl-70, Jo-1, and centromere B) were measured using the autoimmune disease panel on the Bioplex® 2200 (Bio-Rad, Hercules, California) as described49,50,51.
Plasma levels of B lymphocyte stimulator (BLyS; R&D Systems, Minneapolis, MN) and a proliferation-inducing ligand (APRIL; eBioscience/Affymetrix, San Diego, CA) were determined by enzyme-linked immunosorbent assays (ELISA).
Additional analytes including innate and adaptive cytokines, chemokines, and soluble tumor necrosis factor receptor (TNFR) superfamily members were assessed using a custom multiplex panel (ProcartaPlex, Thermo Fisher Scientific/Invitrogen) on the Bioplex 200 Luminex xMAP plate reader (Bio-Rad Technologies, Hercules, CA) as described52,53. Well-specific validity was assessed by AssayCheX™ QC microspheres (Radix BioSolutions, Georgetown, TX, USA), and a standard control serum was included on each plate (Cellgro human AB serum, Cat#2931949, L/N#M1016). Data were acquired on the BioPlex 200 array system (Bio-Rad Technologies, Hercules, CA), with a lower boundary of 100 beads per sample/analyte. Mean inter-assay coefficient of variance (CV) using healthy control serum was 10.5%, similar to other multiplexed cytokine bead-based assays54.
Limit of blank, limit of detection, and limit of quantification were determined using the method of the College of American Pathologists and the Clinical and Laboratory Standards Institute55,56,57,58. The quality control algorithm is detailed in Supplementary Fig. S3. Analytes with >60% undetectable rate were excluded from subsequent analyses. For the 32 analytes passing quality control (Supplementary Table S3), concentrations were interpolated from 5-parameter logistic nonlinear regression standard curves, or assigned a value of 0 if a sample was below the limit of detection.
Immune cell profiling
Immune cells in whole blood were assess by flow cytometry with antibodies for T cell, B cell, and monocyte profiling (Supplementary Methods and Supplementary Table S4). Activated neutrophils and monocytes were identified by expression of anti-activated CD11b (CBRM1/5), which specifically recognizes the CD11b epitope exposed by activation-induced conformational change, above the positive cutoff determined by an isotype control (Supplementary Fig. S4). Debris and doublets were excluded using forward scatter (FSC) and side scatter (SSC). Lymphocytes, monocytes, and granulocytes were roughly separated based on FSC and SSC (Supplementary Fig. S4). Monocytes were further identified by total CD11b expression, and neutrophils were distinguished by CD16 expression. A histogram of fluorescence intensity was used to assess each surface marker. When the histogram showed uni-modal distribution, the median fluorescence intensity was used for further analysis, whereas the 95th percentile was used to define the cut-offs for high expressing populations in multimodal cellular distributions. All flow cytometric results were analyzed by one individual and checked by a second who are both trained in flow cytometry analysis to ensure consistency in data acquisition and analysis.
Statistical analyses
Non-parametric analysis of numerical variables was performed using GraphPad Prism 6.0 (La Jolla, CA). Categorical variables were analyzed using chi-square test, and odds ratios with 95% confidence interval were calculated. False discovery rate (FDR) was used to adjust for multiple testing with the fdr tool R package. A correlation matrix of variables retained after quality control was calculated using R version 3.3.2 and plotted with a p-value filter using corrplot R packages.
Random forest classification was performed in R version 3.3.2 using the randomForest package by Breiman and Cutler, with mtry (number of splitting variables tried at each node) = \(\sqrt{no.variables}\) and ntree (number of decision trees) = 200059. This algorithm generates multiple recursive decision trees based on a randomly selected two-thirds of the samples, then uses the ensemble of decision trees to predict outcomes in the remaining one-third of samples to calculate an internal error, as described60. For the stepwise algorithm, 25 forests were initially used to generate average variable importance with standard deviation based on unscaled accuracy reduction after variable permutation. Noisy variables were excluded based on variable importance, as described61. Briefly, variables were added iteratively in order of importance (from most to least important) to a nested prediction model. Only variables that significantly reduced the prediction error were retained in the final model. Sensitivity, specificity, positive predictive value, and negative predictive value were calculated based on the averaged confusion matrix of 50 forests generated using the best model.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Change history
20 November 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
D’Cruz, D. P., Khamashta, M. A. & Hughes, G. R. Systemic lupus erythematosus. Lancet 369, 587–596, https://doi.org/10.1016/S0140-6736(07)60279-7 (2007).
Yen, E. Y. & Singh, R. R. Brief Report: Lupus-An Unrecognized Leading Cause of Death in Young Females: A Population-Based Study Using Nationwide Death Certificates, 2000-2015. Arthritis Rheumatol 70, 1251–1255, https://doi.org/10.1002/art.40512 (2018).
Petri, M. Disease activity assessment in SLE: do we have the right instruments? Ann Rheum Dis 66(Suppl 3), iii61–64, https://doi.org/10.1136/ard.2007.078477 (2007).
Ruperto, N. et al. International consensus for a definition of disease flare in lupus. Lupus 20, 453–462, https://doi.org/10.1177/0961203310388445 (2011).
Petri, M., Genovese, M., Engle, E. & Hochberg, M. Definition, incidence, and clinical description of flare in systemic lupus erythematosus. A prospective cohort study. Arthritis Rheum 34, 937–944 (1991).
Petri, M., Singh, S., Tesfasyone, H. & Malik, A. Prevalence of flare and influence of demographic and serologic factors on flare risk in systemic lupus erythematosus: a prospective study. J Rheumatol 36, 2476–2480, https://doi.org/10.3899/jrheum.090019 (2009).
van Vollenhoven, R. F. et al. Cumulative Corticosteroid Dose Over Fifty-Two Weeks in Patients With Systemic Lupus Erythematosus: Pooled Analyses From the Phase III Belimumab Trials. Arthritis Rheumatol 68, 2184–2192, https://doi.org/10.1002/art.39682 (2016).
Jacobi, A. M. et al. Correlation between circulating CD27high plasma cells and disease activity in patients with systemic lupus erythematosus. Arthritis Rheum 48, 1332–1342, https://doi.org/10.1002/art.10949 (2003).
Yang, D. H., Chang, D. M., Lai, J. H., Lin, F. H. & Chen, C. H. Significantly higher percentage of circulating CD27(high) plasma cells in systemic lupus erythematosus patients with infection than with disease flare-up. Yonsei Med J 51, 924–931, https://doi.org/10.3349/ymj.2010.51.6.924 (2010).
Petri, M. A. et al. Baseline predictors of systemic lupus erythematosus flares: data from the combined placebo groups in the phase III belimumab trials. Arthritis Rheum 65, 2143–2153, https://doi.org/10.1002/art.37995 (2013).
Ho, A., Magder, L. S., Barr, S. G. & Petri, M. Decreases in anti-double-stranded DNA levels are associated with concurrent flares in patients with systemic lupus erythematosus. Arthritis Rheum 44, 2342–2349 (2001).
Kavanaugh, A. F. & Solomon, D. H. & American College of Rheumatology Ad Hoc Committee on Immunologic Testing, G. Guidelines for immunologic laboratory testing in the rheumatic diseases: anti-DNA antibody tests. Arthritis Rheum 47, 546–555, https://doi.org/10.1002/art.10558 (2002).
ter Borg, E. J., Horst, G., Hummel, E. J., Limburg, P. C. & Kallenberg, C. G. Measurement of increases in anti-double-stranded DNA antibody levels as a predictor of disease exacerbation in systemic lupus erythematosus. A long-term, prospective study. Arthritis Rheum 33, 634–643 (1990).
Froelich, C. J., Wallman, J., Skosey, J. L. & Teodorescu, M. Clinical value of an integrated ELISA system for the detection of 6 autoantibodies (ssDNA, dsDNA, Sm, RNP/Sm, SSA, and SSB). J Rheumatol 17, 192–200 (1990).
Weinstein, A., Bordwell, B., Stone, B., Tibbetts, C. & Rothfield, N. F. Antibodies to native DNA and serum complement (C3) levels. Application to diagnosis and classification of systemic lupus erythematosus. Am J Med 74, 206–216 (1983).
Buyon, J. et al. Reduction in erythrocyte-bound complement activation products and titres of anti-C1q antibodies associate with clinical improvement in systemic lupus erythematosus. Lupus Sci Med 3, e000165, https://doi.org/10.1136/lupus-2016-000165 (2016).
Putterman, C. et al. Cell-bound complement activation products in systemic lupus erythematosus: comparison with anti-double-stranded DNA and standard complement measurements. Lupus Sci Med 1, e000056, https://doi.org/10.1136/lupus-2014-000056 (2014).
Roman, M. J. et al. Prevalence and correlates of accelerated atherosclerosis in systemic lupus erythematosus. N Engl J Med 349, 2399–2406, https://doi.org/10.1056/NEJMoa035471 (2003).
Asherson, R. A. et al. Catastrophic antiphospholipid syndrome: international consensus statement on classification criteria and treatment guidelines. Lupus 12, 530–534 (2003).
Illei, G. G. et al. Combination therapy with pulse cyclophosphamide plus pulse methylprednisolone improves long-term renal outcome without adding toxicity in patients with lupus nephritis. Ann Intern Med 135, 248–257 (2001).
Radis, C. D. et al. Effects of cyclophosphamide on the development of malignancy and on long-term survival of patients with rheumatoid arthritis. A 20-year followup study. Arthritis Rheum 38, 1120–1127 (1995).
Gourley, M. F. et al. Methylprednisolone and cyclophosphamide, alone or in combination, in patients with lupus nephritis. A randomized, controlled trial. Ann Intern Med 125, 549–557 (1996).
Merrill, J. T. et al. The Biomarkers of Lupus Disease Study: A Bold Approach May Mitigate Interference of Background Immunosuppressants in Clinical Trials. Arthritis Rheumatol 69, 1257–1266, https://doi.org/10.1002/art.40086 (2017).
Chaussabel, D. & Baldwin, N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol 14, 271–280, https://doi.org/10.1038/nri3642 (2014).
Chiche, L. et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol 66, 1583–1595, https://doi.org/10.1002/art.38628 (2014).
Uramoto, K. M. et al. Trends in the incidence and mortality of systemic lupus erythematosus, 1950-1992. Arthritis Rheum 42, 46–50, 10.1002/1529-0131(199901)42:1<46::AID-ANR6>3.0.CO;2-2 (1999).
Zhu, T. Y., Tam, L. S., Lee, V. W., Lee, K. K. & Li, E. K. The impact of flare on disease costs of patients with systemic lupus erythematosus. Arthritis Rheum 61, 1159–1167, https://doi.org/10.1002/art.24725 (2009).
Ruiz-Irastorza, G., Egurbide, M. V., Ugalde, J. & Aguirre, C. High impact of antiphospholipid syndrome on irreversible organ damage and survival of patients with systemic lupus erythematosus. Arch Intern Med 164, 77–82, https://doi.org/10.1001/archinte.164.1.77 (2004).
Katsiari, C. G., Liossis, S. N. & Sfikakis, P. P. The pathophysiologic role of monocytes and macrophages in systemic lupus erythematosus: a reappraisal. Semin Arthritis Rheum 39, 491–503, https://doi.org/10.1016/j.semarthrit.2008.11.002 (2010).
Orme, J. & Mohan, C. Macrophages and neutrophils in SLE-An online molecular catalog. Autoimmun Rev 11, 365–372, https://doi.org/10.1016/j.autrev.2011.10.010 (2012).
Buyon, J. P. et al. Surface expression of Gp 165/95, the complement receptor CR3, as a marker of disease activity in systemic Lupus erythematosus. Clin Immunol Immunopathol 46, 141–149 (1988).
Nath, S. K. et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet 40, 152–154, https://doi.org/10.1038/ng.71 (2008).
Kim-Howard, X. et al. ITGAM coding variant (rs1143679) influences the risk of renal disease, discoid rash and immunological manifestations in patients with systemic lupus erythematosus with European ancestry. Ann Rheum Dis 69, 1329–1332, https://doi.org/10.1136/ard.2009.120543 (2010).
Bethunaickan, R. et al. A unique hybrid renal mononuclear phagocyte activation phenotype in murine systemic lupus erythematosus nephritis. J Immunol 186, 4994–5003, https://doi.org/10.4049/jimmunol.1003010 (2011).
Buyon, J. P. et al. Differential phosphorylation of the beta2 integrin CD11b/CD18 in the plasma and specific granule membranes of neutrophils. J Leukoc Biol 61, 313–321 (1997).
Merrill, J. T., Winchester, R. J. & Buyon, J. P. Dynamic state of beta 2 integrin phosphorylation: regulation of neutrophil aggregation involves a phosphatase-dependent pathway. Clin Immunol Immunopathol 71, 216–222 (1994).
Roubey, R. A. et al. Staurosporine inhibits neutrophil phagocytosis but not iC3b binding mediated by CR3 (CD11b/CD18). J Immunol 146, 3557–3562 (1991).
Merrill, J. T., Slade, S. G., Weissmann, G., Winchester, R. & Buyon, J. P. Two pathways of CD11b/CD18-mediated neutrophil aggregation with different involvement of protein kinase C-dependent phosphorylation. J Immunol 145, 2608–2615 (1990).
Chang, N. H. et al. Expanded population of activated antigen-engaged cells within the naive B cell compartment of patients with systemic lupus erythematosus. J Immunol 180, 1276–1284 (2008).
Dolff, S. et al. Peripheral circulating activated b-cell populations are associated with nephritis and disease activity in patients with systemic lupus erythematosus. Scand J Immunol 66, 584–590, https://doi.org/10.1111/j.1365-3083.2007.02008.x (2007).
Guiducci, C. et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature 465, 937–941, https://doi.org/10.1038/nature09102 (2010).
Wang, Z. et al. Role of IFN-gamma in induction of Foxp3 and conversion of CD4+ CD25− T cells to CD4+ Tregs. J Clin Invest 116, 2434–2441, https://doi.org/10.1172/JCI25826 (2006).
Fang, R., Ismail, N., Shelite, T. & Walker, D. H. CD4+ CD25+ Foxp3- T-regulatory cells produce both gamma interferon and interleukin-10 during acute severe murine spotted fever rickettsiosis. Infect Immun 77, 3838–3849, https://doi.org/10.1128/IAI.00349-09 (2009).
Isenberg, D. A. et al. BILAG 2004. Development and initial validation of an updated version of the British Isles Lupus Assessment Group’s disease activity index for patients with systemic lupus erythematosus. Rheumatology (Oxford) 44, 902–906, https://doi.org/10.1093/rheumatology/keh624 (2005).
Yee, C. S. et al. The BILAG-2004 index is sensitive to change for assessment of SLE disease activity. Rheumatology (Oxford) 48, 691–695, https://doi.org/10.1093/rheumatology/kep064 (2009).
Hochberg, M. C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 40, 1725 (1997).
Furie, R. A. et al. Novel evidence-based systemic lupus erythematosus responder index. Arthritis Rheum 61, 1143–1151, https://doi.org/10.1002/art.24698 (2009).
Du, P., Kibbe, W. A. & Lin, S. M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548, https://doi.org/10.1093/bioinformatics/btn224 (2008).
Bruner, B. F. et al. Comparison of autoantibody specificities between traditional and bead-based assays in a large, diverse collection of patients with systemic lupus erythematosus and family members. Arthritis Rheum 64, 3677–3686, https://doi.org/10.1002/art.34651 (2012).
Heinlen, L. D. et al. Clinical criteria for systemic lupus erythematosus precede diagnosis, and associated autoantibodies are present before clinical symptoms. Arthritis Rheum 56, 2344–2351, https://doi.org/10.1002/art.22665 (2007).
Heinlen, L. D. et al. 60 kD Ro and nRNP A frequently initiate human lupus autoimmunity. PLoS One 5, e9599, https://doi.org/10.1371/journal.pone.0009599 (2010).
Lu, R. et al. Dysregulation of innate and adaptive serum mediators precedes systemic lupus erythematosus classification and improves prognostic accuracy of autoantibodies. J Autoimmun 74, 182–193, https://doi.org/10.1016/j.jaut.2016.06.001 (2016).
Munroe, M. E. et al. Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification. Ann Rheum Dis 75, 2014–2021, https://doi.org/10.1136/annrheumdis-2015-208140 (2016).
Munroe, M. E. et al. Proinflammatory adaptive cytokine and shed tumor necrosis factor receptor levels are elevated preceding systemic lupus erythematosus disease flare. Arthritis Rheumatol 66, 1888–1899, https://doi.org/10.1002/art.38573 (2014).
Armbruster, D. A. & Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev 29 Suppl 1, S49–52 (2008).
Lawson, G. M. Defining limit of detection and limit of quantitation as applied to drug of abuse testing: striving for a consensus. Clin Chem 40, 1218–1219 (1994).
(CLSI), C. a. L. S. I. Quantitative Molecular Methods for Infectious Diseases: Approved Guideline - Second Edition, http://shop.clsi.org/site/Sample_pdf/MM06A2_sample.pdf (2010).
Institute, C. a. L. S. in CLSI document (CLSI, 2004).
Shi, T. & Horvath, S. Unsupervised Learning with Random Forest Predictors. Journal of Computational and Graphical Statistics 15, 118–138 (2006).
Breiman, L. Random forests. Machine Learning 45, 5–32, https://doi.org/10.1023/A:1010933404324 (2001).
Genuer, R., Poggi, J. M. & Tuleau-Malot, C. Variable selection using random forests. Pattern Recognition Letters 31, 2225–2236, https://doi.org/10.1016/j.patrec.2010.03.014 (2010).
Acknowledgements
We thank the staff of the Oklahoma Medical Research Foundation (OMRF) Rheumatology Research Clinic for their contributions to the administration and execution of the clinical study, sample acquisition and initial sample processing. We also thank the OMRF Clinical Immunology Laboratory for autoantibody testing and the OMRF Biorepository Core personnel for assistance with sample processing and storage. Research reported in this publication was supported by the National Institute of Allergy and Infectious Disease and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (U19AI082714, P30AR053483, P30AR073750, U01AI101934, and R01AR072401), as well as an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (U54GM104938). The funders had no role in the study design; in the collection, analysis and interpretation of the data; in the writing of the report; nor in the decision to submit the paper for publication. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National institutes of Health.
Author information
Authors and Affiliations
Contributions
R.L., J.M.G., S.S., J.T.M. and J.A.J. designed the study. R.L., J.M.G., S.K., S.R.M., S.S., J.T.M. and J.A.J. acquired data. R.L., J.M.G., H.C., R.L.B., M.E.M., K.B. and J.A.J. analyzed data. All authors assisted with manuscript development and gave approval for publication. J.A.J. had final responsibility for the decision to submit for publication.
Corresponding author
Ethics declarations
Competing Interests
Pfizer supported the BOLD clinical trial as an investigator-initiated study to J.T.M. as the principal investigator. Pfizer had no role in the design of the currently reported biomarker work, in the collection, analysis or interpretation of these biomarker data, or in the writing of this manuscript. S.S. is a former employee of Pfizer. J.T.M. and J.A.J. have received research support from Pfizer, and J.T.M. is a consultant for Pfizer. Patents, with inventors J.A.J. and MEM, have been licensed by OMRF to Progentec Biosciences. All other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lu, R., Guthridge, J.M., Chen, H. et al. Immunologic findings precede rapid lupus flare after transient steroid therapy. Sci Rep 9, 8590 (2019). https://doi.org/10.1038/s41598-019-45135-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-45135-w
This article is cited by
-
An introduction to machine learning and analysis of its use in rheumatic diseases
Nature Reviews Rheumatology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.