Abstract
The giant cell tumor of bone (GCTB) is a locally aggressive primary bone tumor that is composed of mononuclear stroma cells, scattered macrophages, and multinucleated osteoclast-like giant cells which cause pathologic osteolysis. The stroma cells represent the neoplastic population of the tumor and are characterized by the H3F3A mutation G34W. This point mutation is regarded as the driver mutation of GCTB. We have established three new stable H3F3A mutated GCTB cell lines: U-GCT1, U-GCT2, and U-GCT3M. MK-1775 is a Wee1-kinase inhibitor which has been used for blocking of sarcoma growth. In the cell lines we detected Wee1, Cdk1, Cyclin B1, H3K36me3, and Rrm2 as members of the Wee1 pathway. We analyzed the effect of MK-1775 and gemcitabine, alone and in combination, on the growth of the cell lines. The cell lines showed a significant reduction in cell proliferation when treated with MK-1775 or gemcitabine. The combination of both agents led to a further significant reduction in cell proliferation compared to the single agents. Immunohistochemical analysis of 13 GCTB samples revealed that Wee1 and downstream-relevant members are present in GCTB tissue samples. Overall, our work offers valuable new tools for GCTB studies and presents a description of novel biomarkers and molecular targeting strategies.
Similar content being viewed by others
Introduction
The GCTB is a benign but locally aggressive neoplasm. The tumor is predominantly localized in the epiphyseal region of long bones1. Malignant transformation is rare in GCTB occurring in less than 1%, but may rise to 6,6% after radiation therapy2,3. The neoplastic and mitotically active mononuclear stroma cells are characterized by a mutation in H3F3A4. In the presence of macrophage colony–stimulating factor (M-CSF) produced by macrophages the neoplastic cells recruit and activate osteolytic osteoclast giant cells by constitutive expression of receptor activator of nuclear factor kappa-Β ligand (RANKL)1,5,6. Surgery is used as a standard treatment7. However, the tumor recurs in 5 to 25% of patients8. Denosumab is a humanized monoclonal antibody specific for the RANKL and is used in patients with advanced disease9. Recently, several cases of patients with GCTB have been described in which high-grade osteosarcomas arose after treatment with denosumab2,10,11.
The Wee1 pathway has been explored in several types of cancer, such as kidney cancer, colorectal cancer, melanoma, and osteosarcoma12,13,14,15,16. The Wee1 kinase controls the function of Cdk1 by phosphorylation at tyrosine 15, causing an inactivation of Cdk1 (Fig. 1a)17. The active state of Cdk1 forms a complex with Cyclin B1 and drives the cell into mitosis18. Therefore, Wee1 inhibition induces uncontrolled G2/M-transition and may lead to apoptosis through mitotic catastrophe, i.e., due to errors during DNA replication and repair19. The transition into mitosis is further controlled by p53 when cells reach the G2/M checkpoint with damaged DNA or when they are arrested in S-phase due to depletion of the substrates required for DNA synthesis. One mechanism by which p53 blocks cells at the G2/M checkpoint involves the inhibition of CDK120. This suggests that a TP53 mutation sensitizes the cell for WEE1 inhibition13,21. However, further studies showed that the effect is independent of the TP53 status16. In an alternative pathway active Cdk1 mediates phosphorylation of Rrm2, promoting Rrm2 ubiquitylation and degradation, whereas H3K36me3 is present at the promoter of Rrm2 and recruits transcription initiation factors (TAFs). H3F3A mutations in GCTB are known to be associated with an increase in H3K36me322. Severe Rrm2 depletion is thought to lead to dNTP starvation and to induce replication stress. For example, H3K36me3-deficient cell lines, like the kidney carcinoma cell lines A498, have been shown to be selectively killed by MK-1775 via dNTP starvation12. MK-1775, a specific Wee1 inhibitor, has been tested as a possible therapeutic option in sarcomas; e.g., Wee1 inhibition has been shown to sensitize osteosarcoma cells in vitro to chemotherapy or radiation at clinically feasible concentrations15,16,23. Compared to normal tissues, Wee1 is overexpressed in osteosarcomas23. In the breast cancer cell line CAL51, Wee1 is overexpressed and inhibition by MK-1775 is associated with a functional loss of Wee1 leading to cell death underlining the essential role of Wee1 in tumor cell viability24. In a Phase I pharmacological and pharmacodynamics study in patients with melanoma, lung cancer, ovarian cancer, breast cancer or colorectal cancer MK-1775 had a low toxicity profile both as monotherapy and in combination with DNA-damaging agents like gemcitabine (2′,2′-difluoro-2′-deoxycytidine, or dFdC)25. Gemcitabine is a prodrug that is di- or triphosphorylated inside the cell. The triphosphate form (dFdCTP) is a nucleoside analog of cytidine, inhibiting DNA synthesis26. The diphosphate form (dFdCDP) affects the enzyme ribonucleotide reductase and leads to a depletion of deoxycytidine triphosphate (dCTP) pool that potentiates the effects of the drug26,27.
(a) Wee1 inactivates Cdk1 by phosphorylation at tyrosine 1517. Non-phosphorylated Cdk1 forms a complex with Cyclin B1 and induces mitosis18. Non-phosphorylated Cdk1 degrades the ribonucleotide reductase subunit Rrm2. This leads to dNTP starvation and DNA replication stress. H3K36me3 acts as an antagonist promoting Rrm2 transcription. MK-1775, as a Wee1-kinase inhibitor, leads to high Cdk1 activity and uncontrolled G2/M transition. Furthermore, MK-1775 leads to DNA replication stress and to S-phase arrest or apoptosis19,45. p53 protects the cell against DNA replication stress and is a potential inhibitor of Cdk120. Gemcitabine inhibits DNA synthesis as a nucleoside analog of cytidine26.
Based on these data we have investigated the effect of the inhibitor MK-1775 and gemcitabine on the H3F3A-mutated GCTB cell lines. Here, we show that Wee1, Cdk1, H3K36me3, and Rrm2, as crucial players in cell proliferation, are detectable in both GCTB tissue samples and in vitro. We demonstrate that the Wee1 pathway is active in GCTB cell lines and is inhibited by a specific Wee1 inhibitor, MK-1775.
Results
Establishment and characterization of GCTB cell lines
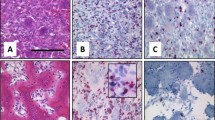
Three cell lines, U-GCT1, U-GCT2, and U-GCT3M (Table 1; Fig. 2a,d,g), were established from H3F3A-mutated GCTBs with typical cytologic morphology of osteoclast-like giant cells and spindle-shaped mononuclear cells (Supplementary Fig. 1a)1. The parental tumors harbored an H3F3A mutation as shown by Sanger sequencing of the relevant exon 2 and immunohistochemistry using a mutation-specific antibody G34W (Supplementary Fig. 1b)28. Short tandem repeats (STR) analysis of the cell lines and the parental tumor confirmed the origin of the cell lines (Supplementary Table 1). De novo DNA sequencing of the established cell lines revealed the H3F3A mutation (Fig. 2b,e,h). The H3F3A mutation was further confirmed by immunochemistry on formalin-fixed and paraffin-embedded cell pellets and Western blot with isolated protein from the cell lines (Figs 2c,f,i and 3b). This proved that the H3F3A-mutated stroma cells had been cultivated4, whereas the osteoclast-like giant cells did not survive further cultivation29. The cell lines have individual doubling times ranging from 7 to 14 days; the cell line established from the lung metastasis has the shortest doubling time of 7 days. The adherent growing cells are spindle-shaped and fibroblastic-like (Fig. 2a,d,g). These characteristics match those of the GCTB cell lines already described30. The TP53 sequence analysis of the appropriate cell line DNA did not identify any relevant mutation in this tumor suppressor gene. The GCTB cell lines and all control cell lines were tested negative for mycoplasma (see Supplementary Fig. 2).
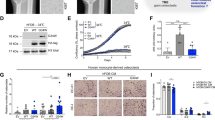
(a) q-PCR determination of the amount of WEE1 expression in the U-GCT cell lines standardized to U-GCT1. (b) Western blot of H3.3 G34W, Wee1, H3K36me3, Rrm2 expression in the three U-GCT cell lines and A498. For the complete Western Blot see Supplementary Fig. 4. (c) Western blot of MK-1775-treated and untreated GCTB cell lines detecting H3K36me3, tyrosine 15 phosphorylated and non-phosphorylated Cdk1. For the complete Western Blot see Supplementary Fig. 5.
Analysis of the Wee1-pathway in GCTB
To obtain an insight into the Wee1 signaling in GCTB, we analyzed the steps of this pathway in the U-GCT cell lines (Fig. 1a). Using quantitative polymerase chain reaction (qPCR) we quantified the expression of WEE1 mRNA in the U-GCT cell lines. U-GCT3M showed the highest WEE1 mRNA levels followed by U-GCT1, whereas U-GCT2 revealed the lowest amount of WEE1 mRNA (Fig. 3a). Wee1, Cdk1, H3K36me3, and Rrm2 were detected consistently in all U-GCT cell lines, whereas A498 was negative for H3K36me3 in Western blot analyses (Fig. 3b,c). Cyclin B1 was not expressed in a detectable amount in any GCTB cell lines by Western blot or immunochemistry, whereas the Jurkat cell line was positive (Fig. 4a,b).
(a) Western blot of Cyclin B1 expression in the three U-GCT cell lines and Jurkat. For the complete Western Blot see Supplementary Fig. 6. (b) Cyclin B1 immunocytological staining with only a few positive cells (see Table 1; sample 11). (c) Cyclin B1 immunocytologic staining of U-GCT1 cell pellets. Cyclin B1. (d) Cyclin B1 immunohistologic staining of placental tissue as positive control.
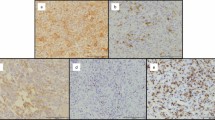
An H3F3A-mutated GCTB cohort of 13 formalin-fixed and paraffin-embedded tissues, including the parental tumors of the U-GCT cell lines, was used to confirm the findings regarding the Wee1 pathway in tissue samples and to determine the proliferation index by Ki-67 (Table 1). Wee1, Cdk1, H3K36me3, and Rrm2 were detected in the tissue samples by immunohistochemistry (Table 1; Supplementary Fig. 3). We found in GCTB tissue samples some very few intermingled Cyclin B1 positive cells with staining of the nucleus and some positive mitotic figures (Fig. 4c), as compared to the positive control (Fig. 4d). The GCTB cohort showed a Ki-67 mean value of about 14.6% and a standard deviation (SD) of 7,8%.
Effect of MK-1775 and gemcitabine on the GCTB cell lines
MTS assays were used to investigate the effect of Wee1 inhibition by MK-1775 on the proliferation of the GCTB cell lines. Again, we used the cell line A498 as control12. The IC50 value of A498 was found to be 77.89 nM (+/−23.84 nM) comparable to published data for the A498 cell line12. Increasing concentrations of MK-1775 led to a reduction in viable cells in all the GCTB cell lines analyzed (Fig. 5a,d,g,j). The IC50 values for these cell lines ranged from 174 to 337.8 nM (Table 2). To determine whether MK-1775 had caused a cytostatic or a cytotoxic effect on the cell lines, we used the apoptosis Western blot cocktail on isolated proteins extracted from untreated and treated cell lines (Fig. 6a). Monitoring apoptosis by analysis of cleaved PARP revealed that cleaved PARP was not detectable in the cell lines analyzed. Moreover, Ki-67 analysis showed a significant reduction in cell proliferation during MK-1775 treatment (Fig. 6b,c,d). Together, these findings argue for a cytostatic rather than a cytotoxic effect of MK-1775. We showed that Wee1 inhibition by MK-1775 leads to a decrease in the amount of H3K36me3 and a reduction in Cdk1 phosphorylation at tyrosine 15 in the GCTB cell lines, whereas Cdk1 levels remained unchanged (Fig. 3c). Treatment with an increasing concentration of gemcitabine led to decreased cell viability of all cell lines (Fig. 5b,e,h,k). The IC50 values for the U-GCTs were between 29.31 and 74.98 nM (Table 2). IC25 values for MK-1775 and gemcitabine were determined and were used either individually or as a combination of both agents (Fig. 5c,f,i,l). The combination led to a significant reduction in cell viability compared to individual treatment. However, we could not detect apoptosis even in the combination of both agents (Fig. 6a).
(a,d,g,j) MTS cell viability test detecting the effect of different MK-1775 concentrations on the various cell lines. (b,e,h,k) MTS cell viability test detecting the effect of different gemcitabine concentrations on the various cell lines. (c,f,i,l) MTS cell viability test detecting the effect of the IC25 single dose of MK-1775, gemcitabine and their combination on the various cell lines.
(a) Western blot analysis for cleaved PARP and Procaspase 3 of U-GCT cell lines as compared to HEK-293 cells (+/− stimulation with bortezomib and TNFα for 24 h). No corresponding bands are apparent in the cell lines after inhibition with 1000 nM MK-1775 or gemcitabine and the combination of both for 72 h. For the complete Western Blot see Supplementary Figs 7 and 8. (b) Box plots of Ki-67 index of cell pellets compared to untreated and MK-1775-treated cell lines. (c,d) Ki-67 immunocytological staining of untreated and MK-1775-treated cells of U-GCT1.
Discussion
In the current study we characterized three newly generated H3F3A mutated GCTB cell lines including the first stable cell line of a GCTB lung metastasis. Since all these cell lines carry the H3F3A mutation, we concluded that these cell lines originate from neoplastic mononuclear stroma cells4. The Wee1 pathway has been analyzed in several tumor types. High Wee1 expression is associated with poor prognosis in pediatric high-grade gliomas, malignant melanoma, and colorectal cancer14,31,32. Wee1 inhibits the G2/M transition through inactivation of Cdk1 by phosphorylation at tyrosine 1517. Consistently, our data also revealed a reduction in Cdk1 phosphorylation at tyrosine 15 in the GCTB cell lines by MK-1775 mediated Wee1 inhibition16,33. MK-1775 was used in several preclinical studies for various cancer types, such as osteosarcoma and colorectal adenocarcinoma. TP53 mutation has been described as a sensitizing factor for inhibition with MK-177513. In line with published data in GCTBs, we found no point mutations in TP5334. Therefore, our results confirm data from osteosarcoma cell lines showing that the efficacy of MK-1775 inhibition is independent of the TP53 mutation status16. Moreover, we show that the GCTBs express Wee1 and other relevant players of this pathway in vitro and in tissue samples. Higher Wee1 expression is associated with higher sensibility for MK-177524. We can confirm this, since UGCT3M shows a significantly higher expression of WEE1 and the lowest IC50 value of the GCTB cell lines. Thus, the inhibitory effect of MK-1775 is linked to the amount of Wee1. A further factor that may influence the Wee1 inhibition is the presence of H3K36me3. H3F3A mutations in GCTB are linked to an increase of H3K36me322, although the functional role of H3K36me3 in GCTBs is unknown. The higher IC50 values for MK-1775-mediated growth inhibition for GCTB cell lines as compared to the H3K36me3-negative cell line A498 is interesting. This observation together with the decrease in H3K36me3 levels upon Wee1 inhibition supports the notion that H3K36me3 has a functional role in this pathway and may have a cell-protecting effect12. Compared with other bone tumors, GCTBs have a lower proliferation rate as detected by a low Ki-67 index35,36. Cyclin B1 acts in a dose-dependent manner. During the S-phase, cyclin B1 increases and forms a complex with Cdk1, leading to mitosis37. These data and the fact that Cyclin B1 in our results across from others is expressed in very low amounts in the GCTBs36 rise the hypothesis that, in our cell system, Wee1 inhibition eventually may cause dNTP starvation rather than an uncontrolled G2/M-transition12. Gemcitabine has been proven to induce growth inhibition in osteosarcomas that were refractory to standard chemotherapy38. Moreover, Wee1 inhibition has been shown to enhance the effect of DNA-damaging agents, such as gemcitabine, in osteosarcomas13,15. We show that the combination of both agents reveals a further significant reduction in cell proliferation compared to the IC25 single application, including in GCTB cells. This combinatorial effect may be explained by the reduced levels of dNTP causing Wee1 inhibition12 and the diminished ribonucleotide reductase activity triggered by dFdCDP, leading to a depletion of the dCTP pool26,27. Equally dFdCTP as a nucleoside analog of cytidine inhibits DNA synthesis26.
First-line treatment for GCTB is surgical curettage of the lesion7. In the event of recurrence or at advanced stages, denosumab as anti-RANK ligand agent has been introduced as a therapeutic concept for GCTB9. The effect of denosumab seems to be limited, since it blocks the osteoclasts and not the neoplastic stroma cells39. In histologic analysis, denosumab-treated GCTBs showed marked giant cell depletion and new bone deposition, leading to substantial histologic overlap with other primary tumors of bone40. Recently, several cases of denosumab-treated GCTBs were described that showed a transformation into a high-grade osteosarcoma with poor prognosis2,10,11. These facts underline the need for novel treatment strategies. In a Phase I study on patients with advanced solid tumors, MK-1775 was well tolerated as monotherapy and in combination with chemotherapeutic substances like gemcitabine25. Since we have detected Wee1, Cdk1, and H3K36me3 in the GCTB tissue samples, these proteins may serve as biomarkers for a definition of patients with GCTB eligible for a potential Wee1 inhibition-based therapy. Taken together, our data argue for an extension of Wee1 therapy to progressed GCTB patients. Therefore, the initiation of a prospective study including GCTB patients expressing the defined biomarkers of the Wee1 pathway by immunohistologic profiling, should now be considered.
Material and Methods
Cell culture and establishment of novel GCTB cell lines
Three novel stable GCTB cell lines were established. U-GCT1 was established from a GCTB measuring 6.0 × 6.0 × 2.0 cm, obtained from the right proximal tibia of a 40-year-old male patient in 05/1999. U-GCT2 was established from a GCTB measuring 4.5 × 4.2 × 2.0 cm obtained from the left proximal tibia of a 17-year-old female patient in 07/2015. The third cell line, U-GCT3M, was established in 03/2016 and obtained from a lung metastasis of a 28-year-old female patient with a GCTB in the left distal femur. The lung manifestation measured 3.0 × 1.3 × 1.0 cm and was localized in segment 6 of the left inferior lobe of the lung. All three patients gave their informed and written consent to the scientific use of their tumor cells. In case of the 17-year-old patient the parental authority additionally gave the informed consent. The research was approved by the local ethics committee of the University of Ulm (reference 369/17 and 372/17).
After extraction, the tumor tissue was prepared for cell culture. The tissue was minced into small pieces and partially digested with collagenase (Sigma-Aldrich, St. Louis, USA, catalog number (cat. #): C9891) 1 mg/ml diluted in 50 nM Trizma Base (Sigma-Aldrich, cat. #: RDD008) and 0.32 nM calcium chloride dihydrate (Sigma-Aldrich, cat. #: C3881), pH 7.4. Isolated cells were cultured in Nunc EasYFlask 25 cm2 cell culture flask (Thermo Scientific, Waltham, USA, cat. #: 156367) with Iscove’s Modified Dulbecco’s Medium with 25 mM HEPES (4:1; Lonza, Basel, Switzerland, cat. #: BE12-726F) with 10% fetal bovine serum (Biochrom AG, Berlin, Germany, cat. #: S0115), 2 mM L-glutamine (Lonza, cat. #: BE17-605E), and penicillin-streptomycin (Lonza, cat. #: DE17-602E). Cells were detached with trypsin (Lonza, cat. #: BE17-161E) before reaching a state of complete confluence. As control cells we used the commercially available cell lines A498, Jurkat, and HEK-293 (Leibniz Institute DSMZ German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). The cell line experiments were performed within 4 months after STR analysis and negative testing for mycoplasma by PCR. All cell lines were cultured at 37 °C in a 5% CO2 incubator applying the same protocol as mentioned above.
Short tandem repeats (STR) analysis
DNA from the cell lines was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany, cat. #: 51304). STR analysis was performed comparing the parental tumors with the corresponding cell lines and genotyping them and the control cell lines by using a standard protocol41.
Polymerase chain reaction (PCR)
Cell lines were tested for the presence of mycoplasma using PCR. PCR amplification was done using specific primers for mycoplasma DNA (primer sequences in Supplementary Table 2). The PCR reaction was carried out in a total volume of 25 µl, including 1 µl DNA or sterile water as a negative control. As positive control isolated DNA from a mycoplasma infected cell line was used. 12,5 µl OneTaq Quick-Load (New England BioLabs, Ipswich, USA, cat. #: #0486S), 1 µl of each primer (5 pmol/µl) and 9, 5 µl of sterile water. PCR was performed using the LabCycler Compact (SensoQuest, Göttingen, Germany, cat. #: 95-3000-048) with a cycling protocol as follows: 10 minutes initial denaturation at 95 °C followed by 55 cycles with 30 seconds at 95 °C, 30 seconds at 50 °C, 30 seconds at 68 °C and final extension at 72 °C for 10 minutes. The PCR products were resolved on a 2% agarose gel.
Gene sequencing
Gene sequencing (Sanger) was performed according to our diagnostic standard protocol28. The tumor tissues and the corresponding cell lines were analyzed for the mutational status of H3F3A. The cell line A498 were used as negative controls. The graphs were generated with FinchTV 1.4.0 (Geospiza Inc., Seattle Washington, USA).
TP53 sequencing analysis
Exons of TP53 were amplified using a Multiplex PCR Kit (Qiagen, cat. #: 206143). Subsequently, PCR products were sequenced using the MiSeq-Sequencer (Illumina, San Diego, USA, cat. #: Y-410-1003) with a coverage of 5000x. The resulting sequences were compared to an hg19 as the reference genome using the MiSeq Reporter Software. The non-synonymous aberration was checked using the Integrated Genome Viewer. Genomic areas were reviewed for clinically relevant TP53-point mutations using http://cancer.sanger.ac.uk/cosmic.
Real-time polymerase chain reaction (qPCR)
RNeasy Mini-Kit (Qiagen, cat. #: 74104) was used for extracting total RNA. A NanoDrop 2000 spectrophotometer (Thermo Scientific, cat. #: ND-2000) was used to control the RNA quality and quantity. Omniscript Reverse Transcription Kit (Qiagen, cat. #: 20511) was used to synthesize single-strand cDNA from total RNA. QuantiTect SYBR Green PCR Kit (Qiagen, cat. #: 204143) and the Light-cycler Rotor Gene Q (Qiagen, cat. #: 9001560) were used to perform the qPCR. The expression of the WEE1 gene was analyzed using the appropriate QuantiTect Primer Assays (Qiagen, cat. #: QT00038199). β-actin and GAPDH served as housekeeping genes (primer sequences in Supplementary Table 2). The PCR reaction was performed as follows: the initial polymerase activation step was 95 °C for 15 minutes, followed by 60 cycles of 94 °C for 15 seconds, at 55 °C (WEE1) or 58 °C (β-Actin and GAPDH) for 30 seconds and at 72 °C for 30 seconds. All experiments were performed in triplicate.
Protein isolation and Western blotting
Protein isolation and Western blots were performed using standard protocols as described elsewhere41. The following primary antibodies were used: R2 (Santa Cruz Biotechnology, Dallas, USA, cat. #: sc-10844, 1:1000), Wee1 (Santa Cruz Biotechnology, cat. #: sc-9037, 1:1000), Anti-Histone H3.3 G34W (RevMAb Biosciences, South San Francisco, USA, cat. #: 31-1145-00, 1:1000), Histone H3K36me3 antibody (Active Motif, Cambridge, UK, cat. #: 61021, 1:1000), Cdc2 p34 (Santa Cruz Biotechnology, cat. #: sc-54, 1:500), Phospho-cdc2 Tyrosine 15 (Cell Signaling, Denver, USA, cat. #: #9111, 1:500), and Cyclin B1 (Santa Cruz Biotechnology, cat. #: sc-245, 1:200). As secondary antibody we used: Anti-Rabbit IgG (Sigma-Aldrich, cat. #: A-9169,1:10000), rabbit anti-goat IgG-HRP (Santa Cruz Biotechnology, cat. #: sc-2768, 1:10000) and goat anti-mouse IgG-HRP (Santa Cruz, cat. #: sc-2005, 1:5000). Anti-β-Actin antibody (Sigma-Aldrich, cat. #: A2228, 1:100000) or Anti-β-Tubulin (Sigma-Aldrich, cat. #: T4026,1:2000) were used to detect β-actin or β-tubulin as a housekeeping protein.
MTS proliferation assay
For inhibition assays, MK-1775 (MedChem Express, Monmouth Junction, USA, cat. #: HY-10993) was diluted in DMSO (Sigma Aldrich, cat. #: D4540) and gemcitabine hydrochloride (Sigma-Aldrich, cat. #: G6423) was diluted in PBS (Lonza, cat. #: 17-516).
For MTS cell proliferation assays, 15000 cells/cm2 of the cell lines were seeded in Nunclon Delta 96-Well MicroWell Plates (Thermo Scientific, cat. #: 163320). An initial incubation at 37 °C and 5% CO2 for 24 hours ensured complete adherence and spreading of the cells. The cells were then incubated for 72 hours under the above-mentioned cell culture conditions at eight different concentrations of the agents increasing in decimal power from 0.01 nM to 100 µM. Untreated cells served as controls. Five wells were prepared for each treatment. As the next step, 10 µl CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS) (Promega, Madison, USA, cat. #: G3580) were added to all wells and the absorption was measured after 3 hours using the Biokinetics Microplate Reader (Bio-Tek Instruments, Winooski, USA, cat. #: EL340) with a 490 nm filter. The results were calibrated in Excel 2016 (Microsoft, Redmond, USA, cat. #: SWR0616). The average of the medium data was subtracted from the other results and the inhibition data were then divided by the average of the dilution solution only-treated cells. The combination effect of MK-1775 and gemcitabine were established by comparing the detected IC25 values of the agents in single-dose and combination as described above.
For apoptosis, Western blot and Ki-67 analysis, the cell lines were cultivated in 2 ml medium in a Nunc EasYFlask 25 cm2 Cell Culture Flask (Thermo Scientific, cat. #: 156367) for 24 hours. The cells were then incubated with 3 mM of the MK-1775, gemcitabine, or both agents in 1 ml medium for 72 h at a concentration of 1 mM. HEK293 cells were treated with 5 ng/ml bortezomib (Santa Cruz, cat. #: sc-217785), diluted in DMSO and 50 ng/ml TNF-α (PeproTech US, Rocky Hill, USA, cat. #: 300-01 A) diluted in medium for 24 hours and were used as positive controls42. Protein isolation and Western blot were conducted using the above-mentioned protocol and subsequently using the primary and secondary antibodies of the apoptosis Western blot cocktail (Abcam, Cambridge, UK, cat. #: ab136812) as previously described. Formalin-fixed and paraffin-embedded cell pellets were prepared using standard protocols. Ki-67 (Dako, Glostrup, Denmark, cat. #: M7240, 1:200) stainings were performed as described below and evaluated on 3 paraffin sections of cell pellet preparations. At least 100 cells were counted on each cytoblock section. Untreated controls were included in every experiment. All experiments were repeated at least three times.
Giant cell tumor tissue bank
GCTB tissue samples with H3F3A mutation from 13 patients were available from the Ulm tissue bank (median age: 40.39 years; range: 17–54 years; 8 males, 5 females). The parental tumors of the cell lines were included (Table 1). Eight patients had localized disease, 4 had recurrences and one patient had metastatic disease. The diagnosis was based on histologic subtyping according to the WHO1. The samples were pseudonymized according to the German law for correct usage of archival tissue for clinical research43. Approval for this procedure was obtained from the local Ethics Committee (vote for usage of archived human material 03/2014) and was in compliance with the ethical principles of the WMA Declaration of Helsinki44.
Immunochemistry
The alkaline phosphatase/RED detection system (Dako, cat. #: K5005) was used for immunohistochemistry and immunochemistry on formalin-fixed and paraffin-embedded tissue or cell pellets via the avidin-biotin-complex-method. The following antibodies were used: Anti-Histone H3.3 G34W (RevMAb Biosciences, cat. #: 31-1145-00, 1:400), Wee1 (Santa Cruz Biotechnology, cat. #: sc-037, 1:25), Cdc2 p34 (Santa Cruz Biotechnology, cat. #: sc-54, 1:200), Cyclin B1 (Santa Cruz Biotechnology, cat. #: sc-245, 1:25), Histone H3K36me3 Antibody (Active Motif, cat. #: 61021, 1:8000), R2 (Santa Cruz Biotechnology, cat. #: sc-10844, 1:100), and Ki-67 Antigen (Dako, cat. #: M7240, 1:200). As retrieval methodes all sides were pretreated by a steamer in EDTA pH 8. Consensus evaluation was done on multihead microscope (CL, TFB, PM).
Statistics
For statistical analysis, two-sided t-tests were performed. A p-value ≤ 0.05% was considered as significant. The graphs were generated with Prism Version 6.01 (Graphpad Software, La Jolla, USA, cat. #: 1820).
All methods were performed in accordance with the relevant guidelines and regulations of good scientific practice of the University of Ulm and the Deutsche Forschungsgemeinschaft (DFG): https://doi.org/10.1002/9783527679188.oth1.
References
Athanasou, N. A., Bansal, M. & Forsyth, R. In WHO Classification of Tumours of Soft Tissue and Bone (eds Fletcher, C. D. M., Bridge, J. A., Hogendoorn, P. C. W. & Mertens, F.) 321–324 (IARC Press, Lyon; France, 2013).
Aponte-Tinao, L. A., Piuzzi, N. S., Roitman, P. & Farfalli, G. L. A High-grade Sarcoma Arising in a Patient With Recurrent Benign Giant Cell Tumor of the Proximal Tibia While Receiving Treatment With Denosumab. Clin. Orthop. Relat. Res. 473, 3050–3055 (2015).
Brien, E. W. et al. Benign giant cell tumor of bone with osteosarcomatous transformation (“dedifferentiated” primary malignant GCT): report of two cases. Skeletal Radiol. 26, 246–255 (1997).
Behjati, S. et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45, 1479–1482 (2013).
Zheng, M. H. et al. The histogenesis of giant cell tumour of bone: a model of interaction between neoplastic cells and osteoclasts. Histol. Histopathol. 16, 297–307 (2001).
Roux, S. et al. RANK (receptor activator of nuclear factor kappa B) and RANK ligand are expressed in giant cell tumors of bone. Am. J. Clin. Pathol. 117, 210–216 (2002).
Sobti, A., Agrawal, P., Agarwala, S. & Agarwal, M. Giant Cell Tumor of Bone - An Overview. Arch. Bone Jt. Surg. 4, 2–9 (2016).
Klenke, F. M., Wenger, D. E., Inwards, C. Y., Rose, P. S. & Sim, F. H. Giant cell tumor of bone: risk factors for recurrence. Clin. Orthop. Relat. Res. 469, 591–599 (2011).
Chawla, S. et al. Safety and efficacy of denosumab for adults and skeletally mature adolescents with giant cell tumour of bone: interim analysis of an open-label, parallel-group, phase 2 study. The Lancet Oncology 14, 901–908 (2013).
Broehm, C. J., Garbrecht, E. L., Wood, J. & Bocklage, T. Two Cases of Sarcoma Arising in Giant Cell Tumor of Bone Treated with Denosumab. Case Rep. Med. 2015, 6 (2015).
Tsukamoto, S. et al. Development of high-grade osteosarcoma in a patient with recurrent giant cell tumor of the ischium while receiving treatment with denosumab. Jpn J Clin Oncol. 47(11), 1090–1096 (2017).
Pfister, S. X. et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer. Cell. 28, 557–568 (2015).
Hirai, H. et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer. Ther. 8, 2992–3000 (2009).
Magnussen, G. I. et al. High expression of Wee1 is associated with poor disease-free survival in malignant melanoma: potential for targeted therapy. PLoS One 7, e38254 (2012).
Kreahling, J. M. et al. Wee1 inhibition by MK-1775 leads to tumor inhibition and enhances efficacy of gemcitabine in human sarcomas. PLoS One 8, e57523 (2013).
Kreahling, J. M. et al. MK1775, a selective Wee1 inhibitor, shows single-agent antitumor activity against sarcoma cells. Mol. Cancer. Ther. 11, 174–182 (2012).
Heald, R., McLoughlin, M. & McKeon, F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell 74, 463–474 (1993).
Smits, V. A. & Medema, R. H. Checking out the G(2)/M transition. Biochim. Biophys. Acta 1519, 1–12 (2001).
Hamer, D. W. et al. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin. Cancer Res. 17, 4200–4207 (2011).
Taylor, W. R. & Stark, G. R. Regulation of the G2/M transition by p53. Oncogene 20, 1803–1815 (2001).
Leijen, S., Beijnen, J. H. & Schellens, J. H. Abrogation of the G2 checkpoint by inhibition of Wee-1 kinase results in sensitization of p53-deficient tumor cells to DNA-damaging agents. Curr. Clin. Pharmacol. 5, 186–191 (2010).
Cleven, A. H. et al. Mutation Analysis of H3F3A and H3F3B as a Diagnostic Tool for Giant Cell Tumor of Bone and Chondroblastoma. Am. J. Surg. Pathol. 39, 1576–1583 (2015).
PosthumaDeBoer, J. et al. WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer 11, 156 (2011).
Iorns, E. et al. Integrated functional, gene expression and genomic analysis for the identification of cancer targets. PLoS One 4, e5120 (2009).
Leijen, S. et al. A phase I pharmacological and pharmacodynamic study of MK-1775, a Wee1 tyrosine kinase inhibitor, in monotherapy and combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. JCO 28, 3067–3067 (2010).
Plunkett, W. et al. 2′,2′-Difluorodeoxycytidine Metabolism and Mechanism of Action In Human Leukemia Cells. Nucleosides and Nucleotides 8, 775–785 (1989).
Heinemann, V. et al. Inhibition of ribonucleotide reduction in CCRF-CEM cells by 2′,2′-difluorodeoxycytidine. Mol. Pharmacol. 38, 567–572 (1990).
Lüke, J. et al. H3F3A mutation in giant cell tumour of the bone is detected by immunohistochemistry using a monoclonal antibody against the G34W mutated site of the histone H3.3 variant. Histopathology 71, 125–133 (2017).
Atkins, G. J. et al. Expression of osteoclast differentiation signals by stromal elements of giant cell tumors. J. Bone Miner. Res. 15, 640–649 (2000).
Liu, L. et al. Enrichment of c-Met+ tumorigenic stromal cells of giant cell tumor of bone and targeting by cabozantinib. Cell. Death Dis. 5, e1471 (2014).
Mueller, S. et al. Targeting Wee1 for the treatment of pediatric high-grade gliomas. Neuro Oncol. 16, 352–360 (2014).
Ge, X.- et al. Upregulation of WEE1 is a potential prognostic biomarker for patients with colorectal cancer. Oncology Letters 13(6), 4341–4348 (2017).
Kiviharju-af Hallstrom, T. M. et al. Human prostate epithelium lacks Wee1A-mediated DNA damage-induced checkpoint enforcement. Proc. Natl. Acad. Sci. USA 104, 7211–7216 (2007).
Moskovszky, L. et al. Genomic instability in giant cell tumor of bone. A study of 52 cases using DNA ploidy, relocalization FISH, and array-CGH analysis. Genes Chromosomes Cancer 48, 468–479 (2009).
Scotlandi, K. et al. Clinical relevance of Ki-67 expression in bone tumors. Cancer 75, 806–814 (1995).
Kauzman, A. et al. Cyclin alterations in giant cell tumor of bone. Mod. Pathol. 16, 210–218 (2003).
Jin, P., Hardy, S. & Morgan, D. O. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J. Cell Biol. 141, 875–885 (1998).
Merimsky, O. et al. Gemcitabine in soft tissue or bone sarcoma resistant to standard chemotherapy: a phase II study. Cancer Chemother. Pharmacol. 45, 177–181 (2000).
Balke, M. et al. A short-term in vivo model for giant cell tumor of bone. BMC Cancer 11, 241–241 (2011).
Wojcik, J. et al. Denosumab-treated Giant Cell Tumor of Bone Exhibits Morphologic Overlap With Malignant Giant Cell Tumor of Bone. Am. J. Surg. Pathol. 40, 72–80 (2016).
Jager, D. et al. HOXA7, HOXA9, and HOXA10 are differentially expressed in clival and sacral chordomas. Sci. Rep. 7, 2032-017–02174-5 (2017).
An, J. et al. Drug interactions between the proteasome inhibitor bortezomib and cytotoxic chemotherapy, tumor necrosis factor (TNF) alpha, and TNF-related apoptosis-inducing ligand in prostate cancer. Clin. Cancer Res. 9, 4537–4545 (2003).
Zentrale Ethikkommission bei der Bundesärztekammer. Mitteilungen: Die (Weiter-)Verwendung von menschlichen Körpermaterialien für Zwecke medizinischer Forschung. Dtsch Ärztebl International, A-1632 (2003).
World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 310, 2191–2194 (2013).
Dixon, H. & Norbury, C. J. Therapeutic exploitation of checkpoint defects in cancer cells lacking p53 function. Cell. Cycle 1, 362–368 (2002).
Acknowledgements
The technical help of Michaela Buck, Karola Dorsch, Julian Giesche, Daniela Jäger, Elena Kelsch, Ulrike Kostezka, André Lechel, Judith Modeste, Elena Moser, Juliane Nell, Nina Ushmorova, and Malena Zahn is greatly acknowledged. The International Graduate School in Molecular Medicine Ulm (IGradU) supported Christoph Lübbehüsen with a doctoral scholarship.
Author information
Authors and Affiliations
Contributions
C.L., J.L., K.M., R.M., T.F.E.B. and P.M. conceived the experiments and analysed the data. C.L. and C.S. conducted the experiments. C.L., A.v.B. and M.S. collected material and primary data. C.L., K.M. and T.F.E.B. wrote the manuscript. C.L. and K.M., designed the figures and tables. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lübbehüsen, C., Lüke, J., Seeling, C. et al. Characterization of Three Novel H3F3A-mutated Giant Cell Tumor Cell Lines and Targeting of Their Wee1 Pathway. Sci Rep 9, 6458 (2019). https://doi.org/10.1038/s41598-019-42611-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-42611-1
This article is cited by
-
Der Riesenzelltumor des Knochens – ein Update
Die Pathologie (2023)
-
Profiling of three H3F3A-mutated and denosumab-treated giant cell tumors of bone points to diverging pathways during progression and malignant transformation
Scientific Reports (2021)
-
Mutation-driven epigenetic alterations as a defining hallmark of central cartilaginous tumours, giant cell tumour of bone and chondroblastoma
Virchows Archiv (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.